Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
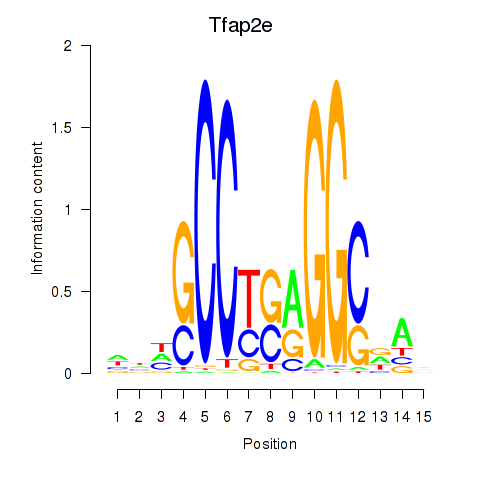
Results for Tfap2e
Z-value: 0.71
Transcription factors associated with Tfap2e
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2e
|
ENSMUSG00000042477.7 | transcription factor AP-2, epsilon |
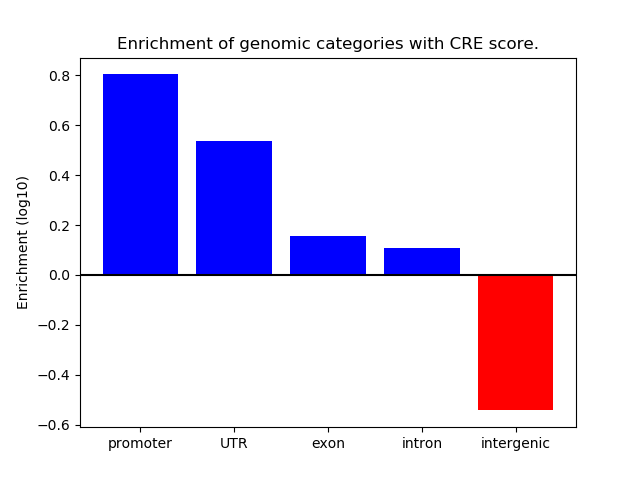
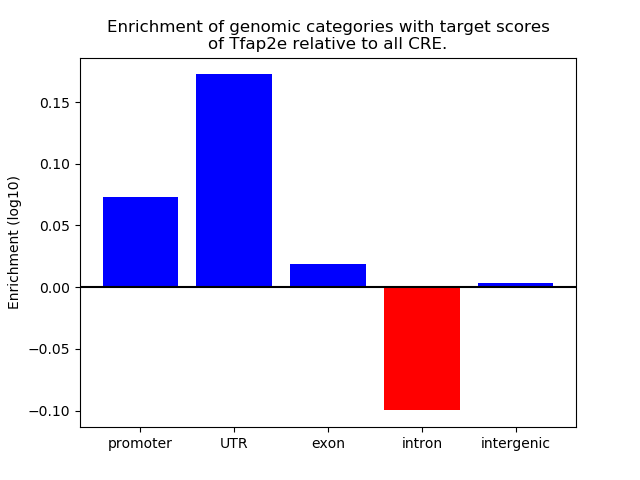
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_126740288_126740466 | Tfap2e | 4137 | 0.156133 | 0.34 | 8.4e-03 | Click! |
| chr4_126735142_126736537 | Tfap2e | 401 | 0.792028 | -0.33 | 9.1e-03 | Click! |
| chr4_126733989_126734295 | Tfap2e | 2098 | 0.232339 | 0.21 | 1.0e-01 | Click! |
| chr4_126734304_126734958 | Tfap2e | 1609 | 0.289741 | 0.21 | 1.1e-01 | Click! |
| chr4_126733832_126733983 | Tfap2e | 2333 | 0.214920 | 0.15 | 2.5e-01 | Click! |
Activity of the Tfap2e motif across conditions
Conditions sorted by the z-value of the Tfap2e motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
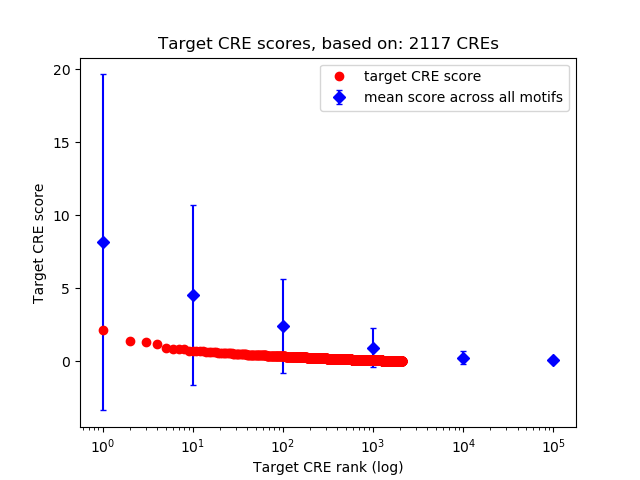
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_162220626_162221297 | 2.15 |
Dnm3os |
dynamin 3, opposite strand |
1093 |
0.42 |
| chr12_73042017_73042233 | 1.38 |
Six1 |
sine oculis-related homeobox 1 |
690 |
0.71 |
| chr10_3863416_3864702 | 1.31 |
Gm16149 |
predicted gene 16149 |
5548 |
0.21 |
| chr12_33955777_33956381 | 1.15 |
Twist1 |
twist basic helix-loop-helix transcription factor 1 |
1592 |
0.42 |
| chr3_86541860_86542342 | 0.88 |
Lrba |
LPS-responsive beige-like anchor |
68 |
0.98 |
| chr8_50916657_50916808 | 0.87 |
C130073E24Rik |
RIKEN cDNA C130073E24 gene |
265 |
0.96 |
| chr1_172484027_172485046 | 0.85 |
Igsf9 |
immunoglobulin superfamily, member 9 |
2222 |
0.17 |
| chr11_101815690_101815909 | 0.84 |
Etv4 |
ets variant 4 |
30428 |
0.12 |
| chr18_60646910_60648302 | 0.71 |
Synpo |
synaptopodin |
666 |
0.69 |
| chr8_12399326_12400483 | 0.70 |
Gm25239 |
predicted gene, 25239 |
3501 |
0.16 |
| chr18_79105943_79106581 | 0.69 |
Setbp1 |
SET binding protein 1 |
3129 |
0.37 |
| chr5_115438447_115439259 | 0.68 |
4930430O22Rik |
RIKEN cDNA 4930430O22 gene |
2209 |
0.13 |
| chr11_79661785_79661936 | 0.68 |
Rab11fip4 |
RAB11 family interacting protein 4 (class II) |
1298 |
0.3 |
| chr11_85833878_85836704 | 0.65 |
Tbx2 |
T-box 2 |
2740 |
0.17 |
| chr2_30554051_30554283 | 0.64 |
Gm14487 |
predicted gene 14487 |
19438 |
0.15 |
| chr18_77563866_77564228 | 0.63 |
Rnf165 |
ring finger protein 165 |
562 |
0.8 |
| chr16_93558150_93558964 | 0.61 |
Setd4 |
SET domain containing 4 |
25451 |
0.12 |
| chr14_118231082_118232196 | 0.61 |
Gm4675 |
predicted gene 4675 |
4593 |
0.14 |
| chr6_6863513_6864355 | 0.60 |
Dlx6 |
distal-less homeobox 6 |
136 |
0.91 |
| chr6_112945034_112945954 | 0.57 |
Srgap3 |
SLIT-ROBO Rho GTPase activating protein 3 |
1260 |
0.31 |
| chr12_9577276_9578110 | 0.57 |
Osr1 |
odd-skipped related transcription factor 1 |
3252 |
0.23 |
| chr10_97566192_97567945 | 0.57 |
Lum |
lumican |
1940 |
0.32 |
| chr8_75113324_75113614 | 0.57 |
Mcm5 |
minichromosome maintenance complex component 5 |
3845 |
0.17 |
| chr9_41378412_41379411 | 0.56 |
Mir100hg |
Mir100 Mirlet7a-2 Mir125b-1 cluster host gene |
2350 |
0.27 |
| chr11_97626854_97627270 | 0.55 |
Epop |
elongin BC and polycomb repressive complex 2 associated protein |
2640 |
0.14 |
| chr6_53267728_53268574 | 0.54 |
Creb5 |
cAMP responsive element binding protein 5 |
19119 |
0.22 |
| chr17_24521651_24521802 | 0.53 |
Traf7 |
TNF receptor-associated factor 7 |
2518 |
0.1 |
| chr14_31124934_31125916 | 0.52 |
Smim4 |
small integral membrane protein 4 |
3413 |
0.14 |
| chr4_139142908_139143066 | 0.52 |
Hspe1-ps4 |
heat shock protein 1 (chaperonin 10), pseudogene 4 |
7210 |
0.14 |
| chr14_44858459_44859503 | 0.52 |
Gm16976 |
predicted gene, 16976 |
27 |
0.85 |
| chr6_77244137_77244322 | 0.51 |
Lrrtm1 |
leucine rich repeat transmembrane neuronal 1 |
1307 |
0.55 |
| chr6_30709196_30709388 | 0.51 |
Gm14528 |
predicted gene 14528 |
10581 |
0.11 |
| chr1_133187933_133188192 | 0.51 |
Plekha6 |
pleckstrin homology domain containing, family A member 6 |
6741 |
0.19 |
| chr15_72033547_72034526 | 0.50 |
Col22a1 |
collagen, type XXII, alpha 1 |
191 |
0.97 |
| chr14_54573090_54573690 | 0.48 |
Ajuba |
ajuba LIM protein |
1308 |
0.24 |
| chr2_39003642_39003858 | 0.48 |
Rpl35 |
ribosomal protein L35 |
1293 |
0.23 |
| chr12_111424483_111424634 | 0.48 |
Exoc3l4 |
exocyst complex component 3-like 4 |
2997 |
0.16 |
| chr14_122475443_122476757 | 0.48 |
Zic2 |
zinc finger protein of the cerebellum 2 |
665 |
0.44 |
| chr2_73356737_73357357 | 0.47 |
Gm13703 |
predicted gene 13703 |
221 |
0.89 |
| chr2_167540595_167541629 | 0.46 |
Snai1 |
snail family zinc finger 1 |
2917 |
0.16 |
| chr9_98958573_98959491 | 0.46 |
Foxl2os |
forkhead box L2, opposite strand |
3733 |
0.14 |
| chr3_95931539_95931909 | 0.46 |
Anp32e |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
1077 |
0.31 |
| chr9_91404809_91406365 | 0.45 |
Gm29478 |
predicted gene 29478 |
1113 |
0.42 |
| chr15_95527528_95528774 | 0.45 |
Nell2 |
NEL-like 2 |
27 |
0.99 |
| chr12_108958975_108959883 | 0.44 |
Gm47980 |
predicted gene, 47980 |
1483 |
0.33 |
| chr6_100422018_100422496 | 0.44 |
Gm23234 |
predicted gene, 23234 |
72557 |
0.09 |
| chr17_34325316_34325467 | 0.44 |
H2-Eb2 |
histocompatibility 2, class II antigen E beta2 |
274 |
0.8 |
| chr12_108179845_108180611 | 0.44 |
Ccnk |
cyclin K |
490 |
0.66 |
| chr1_121527427_121528842 | 0.43 |
Htr5b |
5-hydroxytryptamine (serotonin) receptor 5B |
331 |
0.84 |
| chr5_111414297_111414695 | 0.43 |
Mn1 |
meningioma 1 |
2866 |
0.22 |
| chr10_45783632_45783865 | 0.43 |
D030045P18Rik |
RIKEN cDNA D030045P18 gene |
23814 |
0.18 |
| chr9_61163718_61163950 | 0.43 |
I730028E13Rik |
RIKEN cDNA I730028E13 gene |
25319 |
0.14 |
| chr17_29326157_29327438 | 0.43 |
Gm46603 |
predicted gene, 46603 |
4248 |
0.12 |
| chr7_25291976_25292552 | 0.43 |
Cic |
capicua transcriptional repressor |
329 |
0.76 |
| chr14_40894027_40894178 | 0.43 |
Sh2d4b |
SH2 domain containing 4B |
836 |
0.67 |
| chr17_49816188_49816339 | 0.43 |
Gm16555 |
predicted gene 16555 |
5562 |
0.26 |
| chr13_43210781_43210932 | 0.42 |
Tbc1d7 |
TBC1 domain family, member 7 |
39355 |
0.15 |
| chr4_139825248_139825758 | 0.42 |
Pax7 |
paired box 7 |
7504 |
0.23 |
| chr11_60294712_60295354 | 0.42 |
Gm27711 |
predicted gene, 27711 |
7791 |
0.14 |
| chr3_89435022_89435319 | 0.42 |
Pbxip1 |
pre B cell leukemia transcription factor interacting protein 1 |
1536 |
0.19 |
| chr8_122615874_122616025 | 0.41 |
Trappc2l |
trafficking protein particle complex 2-like |
4303 |
0.1 |
| chr7_101833612_101833784 | 0.41 |
Gm10602 |
predicted gene 10602 |
2805 |
0.12 |
| chr7_4747695_4747846 | 0.41 |
Kmt5c |
lysine methyltransferase 5C |
1962 |
0.14 |
| chr2_32307829_32308017 | 0.40 |
Dnm1 |
dynamin 1 |
2712 |
0.12 |
| chr13_15464574_15464999 | 0.40 |
Gli3 |
GLI-Kruppel family member GLI3 |
806 |
0.46 |
| chr6_42265062_42266248 | 0.39 |
Casp2 |
caspase 2 |
535 |
0.62 |
| chr5_137043355_137043506 | 0.39 |
Ap1s1 |
adaptor protein complex AP-1, sigma 1 |
1977 |
0.19 |
| chr5_24321335_24322296 | 0.39 |
Kcnh2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
439 |
0.73 |
| chr11_53637044_53637520 | 0.39 |
Il13 |
interleukin 13 |
2580 |
0.17 |
| chr7_28386729_28387087 | 0.39 |
Med29 |
mediator complex subunit 29 |
5800 |
0.08 |
| chr4_19871659_19871930 | 0.38 |
Atp6v0d2 |
ATPase, H+ transporting, lysosomal V0 subunit D2 |
45146 |
0.14 |
| chr15_100644899_100645050 | 0.38 |
Mir6961 |
microRNA 6961 |
4255 |
0.1 |
| chr17_37196656_37197851 | 0.38 |
Olfr94 |
olfactory receptor 94 |
758 |
0.35 |
| chr5_136860922_136861924 | 0.38 |
Gm20485 |
predicted gene 20485 |
8299 |
0.14 |
| chr9_61530173_61530451 | 0.38 |
Gm34424 |
predicted gene, 34424 |
3103 |
0.31 |
| chr11_119514733_119515649 | 0.38 |
Endov |
endonuclease V |
13213 |
0.13 |
| chr17_17408831_17408982 | 0.38 |
Gm26873 |
predicted gene, 26873 |
3087 |
0.2 |
| chr13_78205305_78205829 | 0.38 |
A830082K12Rik |
RIKEN cDNA A830082K12 gene |
2780 |
0.19 |
| chr2_174760566_174761903 | 0.37 |
Edn3 |
endothelin 3 |
182 |
0.96 |
| chr11_51853445_51853596 | 0.37 |
Jade2 |
jade family PHD finger 2 |
3605 |
0.22 |
| chr9_69469003_69469456 | 0.37 |
Anxa2 |
annexin A2 |
9236 |
0.12 |
| chr8_123416536_123417431 | 0.36 |
Tubb3 |
tubulin, beta 3 class III |
5393 |
0.08 |
| chr13_53469792_53470715 | 0.36 |
Msx2 |
msh homeobox 2 |
2821 |
0.27 |
| chr10_8813926_8814547 | 0.36 |
Gm25410 |
predicted gene, 25410 |
9911 |
0.2 |
| chr4_148037756_148037907 | 0.36 |
Clcn6 |
chloride channel, voltage-sensitive 6 |
731 |
0.4 |
| chr6_145075061_145076261 | 0.36 |
Bcat1 |
branched chain aminotransferase 1, cytosolic |
523 |
0.72 |
| chr2_167559853_167560205 | 0.36 |
Gm11476 |
predicted gene 11476 |
6715 |
0.12 |
| chrX_49484933_49485344 | 0.36 |
Arhgap36 |
Rho GTPase activating protein 36 |
537 |
0.82 |
| chr4_11123032_11124084 | 0.36 |
Gm11827 |
predicted gene 11827 |
400 |
0.77 |
| chr11_60701603_60702274 | 0.36 |
Llgl1 |
LLGL1 scribble cell polarity complex component |
2093 |
0.12 |
| chr2_93649530_93649681 | 0.35 |
Alx4 |
aristaless-like homeobox 4 |
7217 |
0.25 |
| chr4_32923038_32923201 | 0.35 |
Ankrd6 |
ankyrin repeat domain 6 |
336 |
0.87 |
| chr9_18292786_18293406 | 0.35 |
Chordc1 |
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
708 |
0.62 |
| chr17_24765085_24765600 | 0.35 |
Hs3st6 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
12326 |
0.07 |
| chr3_122518339_122518794 | 0.34 |
Bcar3 |
breast cancer anti-estrogen resistance 3 |
6080 |
0.14 |
| chr12_79924950_79925998 | 0.34 |
9430078K24Rik |
RIKEN cDNA 9430078K24 gene |
20 |
0.98 |
| chr4_95153802_95153953 | 0.34 |
Gm28096 |
predicted gene 28096 |
2137 |
0.28 |
| chr14_122477033_122477655 | 0.34 |
Zic2 |
zinc finger protein of the cerebellum 2 |
756 |
0.48 |
| chr15_10711072_10711949 | 0.34 |
Rai14 |
retinoic acid induced 14 |
2030 |
0.32 |
| chr10_61423139_61423290 | 0.34 |
Nodal |
nodal |
5242 |
0.12 |
| chr9_40804526_40806209 | 0.33 |
Snord14e |
small nucleolar RNA, C/D box 14E |
619 |
0.38 |
| chrX_99819606_99819757 | 0.33 |
Tmem28 |
transmembrane protein 28 |
1340 |
0.52 |
| chr8_95084589_95084801 | 0.33 |
Katnb1 |
katanin p80 (WD40-containing) subunit B 1 |
2099 |
0.17 |
| chr11_98922257_98922979 | 0.33 |
Cdc6 |
cell division cycle 6 |
2152 |
0.18 |
| chr10_96955242_96955756 | 0.33 |
Gm33981 |
predicted gene, 33981 |
3372 |
0.31 |
| chr8_25070198_25070900 | 0.33 |
Plekha2 |
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
20797 |
0.11 |
| chr4_130544569_130544756 | 0.33 |
Nkain1 |
Na+/K+ transporting ATPase interacting 1 |
13326 |
0.24 |
| chr2_157341643_157341794 | 0.32 |
Ghrh |
growth hormone releasing hormone |
4292 |
0.19 |
| chr15_99601478_99601706 | 0.32 |
Aqp6 |
aquaporin 6 |
192 |
0.89 |
| chr10_116760729_116761294 | 0.32 |
4930579P08Rik |
RIKEN cDNA 4930579P08 gene |
31758 |
0.14 |
| chr3_99261050_99262399 | 0.32 |
Gm43120 |
predicted gene 43120 |
439 |
0.81 |
| chr7_16611263_16612711 | 0.32 |
Gm29443 |
predicted gene 29443 |
1837 |
0.17 |
| chr5_92998100_92998342 | 0.32 |
Gm6450 |
predicted gene 6450 |
5610 |
0.16 |
| chr18_34933820_34933974 | 0.32 |
Etf1 |
eukaryotic translation termination factor 1 |
1890 |
0.22 |
| chr2_84741934_84742491 | 0.32 |
Mir130a |
microRNA 130a |
1034 |
0.27 |
| chr2_92600016_92600773 | 0.32 |
Chst1 |
carbohydrate sulfotransferase 1 |
687 |
0.61 |
| chr8_48614074_48614225 | 0.32 |
Gm45772 |
predicted gene 45772 |
17020 |
0.26 |
| chr19_21785153_21785677 | 0.32 |
Cemip2 |
cell migration inducing hyaluronidase 2 |
7027 |
0.22 |
| chr17_25556052_25557244 | 0.32 |
Gm50026 |
predicted gene, 50026 |
4565 |
0.09 |
| chr5_111418246_111419747 | 0.31 |
Mn1 |
meningioma 1 |
1554 |
0.34 |
| chr13_110280269_110281178 | 0.31 |
Rab3c |
RAB3C, member RAS oncogene family |
178 |
0.96 |
| chr12_100998948_100999342 | 0.31 |
Gm36756 |
predicted gene, 36756 |
6173 |
0.13 |
| chr19_45795603_45796547 | 0.31 |
Kcnip2 |
Kv channel-interacting protein 2 |
371 |
0.82 |
| chr9_22005805_22006085 | 0.31 |
Prkcsh |
protein kinase C substrate 80K-H |
377 |
0.71 |
| chr17_25935201_25935431 | 0.31 |
Pigq |
phosphatidylinositol glycan anchor biosynthesis, class Q |
249 |
0.79 |
| chr6_92786590_92787798 | 0.31 |
A730049H05Rik |
RIKEN cDNA A730049H05 gene |
29284 |
0.19 |
| chr15_103334436_103334587 | 0.31 |
Zfp385a |
zinc finger protein 385A |
184 |
0.9 |
| chr15_103302622_103303166 | 0.31 |
Gm49482 |
predicted gene, 49482 |
1829 |
0.19 |
| chr11_60127277_60127428 | 0.30 |
4930412M03Rik |
RIKEN cDNA 4930412M03 gene |
12122 |
0.14 |
| chr11_76577733_76577884 | 0.30 |
Abr |
active BCR-related gene |
81 |
0.97 |
| chr10_57040338_57040864 | 0.30 |
Gm36827 |
predicted gene, 36827 |
58369 |
0.13 |
| chr9_44381420_44381612 | 0.30 |
Hyou1 |
hypoxia up-regulated 1 |
987 |
0.22 |
| chr7_28151731_28151882 | 0.30 |
Gm31024 |
predicted gene, 31024 |
17821 |
0.09 |
| chr17_46618185_46620270 | 0.30 |
Ptk7 |
PTK7 protein tyrosine kinase 7 |
10277 |
0.1 |
| chr9_98668581_98668734 | 0.30 |
Pisrt1 |
polled intersex syndrome regulated transcript 1 |
37609 |
0.12 |
| chr4_150871780_150871938 | 0.30 |
Errfi1 |
ERBB receptor feedback inhibitor 1 |
16786 |
0.12 |
| chr10_115478421_115479410 | 0.29 |
Lgr5 |
leucine rich repeat containing G protein coupled receptor 5 |
15836 |
0.19 |
| chr3_99248252_99249856 | 0.29 |
Tbx15 |
T-box 15 |
4706 |
0.18 |
| chr6_79538142_79538293 | 0.29 |
Gm6261 |
predicted gene 6261 |
2991 |
0.4 |
| chr5_147108738_147108889 | 0.29 |
Polr1d |
polymerase (RNA) I polypeptide D |
3447 |
0.2 |
| chr2_30687005_30687641 | 0.29 |
Gm14486 |
predicted gene 14486 |
9312 |
0.15 |
| chr12_112143792_112144988 | 0.29 |
Kif26a |
kinesin family member 26A |
1818 |
0.26 |
| chr19_7069218_7069544 | 0.29 |
Macrod1 |
mono-ADP ribosylhydrolase 1 |
12571 |
0.1 |
| chr9_27001001_27001320 | 0.29 |
Thyn1 |
thymocyte nuclear protein 1 |
1432 |
0.26 |
| chr11_120231879_120233008 | 0.29 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
504 |
0.52 |
| chr7_68369791_68370269 | 0.29 |
Gm44889 |
predicted gene 44889 |
14749 |
0.12 |
| chr5_35388341_35389287 | 0.29 |
Hmx1 |
H6 homeobox 1 |
294 |
0.51 |
| chr12_72941105_72942093 | 0.29 |
4930447C04Rik |
RIKEN cDNA 4930447C04 gene |
825 |
0.49 |
| chr9_69669605_69671558 | 0.29 |
Gm47203 |
predicted gene, 47203 |
70827 |
0.09 |
| chr1_194208152_194208457 | 0.28 |
4930503O07Rik |
RIKEN cDNA 4930503O07 gene |
14455 |
0.3 |
| chr17_29794989_29795742 | 0.28 |
Gm50016 |
predicted gene, 50016 |
24022 |
0.14 |
| chr8_120617013_120617164 | 0.28 |
1190005I06Rik |
RIKEN cDNA 1190005I06 gene |
17275 |
0.08 |
| chrX_134405492_134405850 | 0.28 |
Drp2 |
dystrophin related protein 2 |
869 |
0.59 |
| chr11_116515408_116515996 | 0.28 |
Rpl36-ps1 |
ribosomal protein L36, pseudogene 1 |
11166 |
0.09 |
| chr14_55833919_55834070 | 0.28 |
Nfatc4 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 4 |
3725 |
0.11 |
| chr7_144285786_144285937 | 0.28 |
Shank2 |
SH3 and multiple ankyrin repeat domains 2 |
1316 |
0.55 |
| chr2_119196463_119196630 | 0.28 |
Gm14138 |
predicted gene 14138 |
9029 |
0.09 |
| chr5_75158634_75158785 | 0.28 |
Pdgfra |
platelet derived growth factor receptor, alpha polypeptide |
1876 |
0.2 |
| chr11_88584739_88584890 | 0.27 |
Msi2 |
musashi RNA-binding protein 2 |
5333 |
0.28 |
| chr11_83300807_83300958 | 0.27 |
Ap2b1 |
adaptor-related protein complex 2, beta 1 subunit |
1601 |
0.18 |
| chr6_52231944_52233433 | 0.27 |
Hoxa10 |
homeobox A10 |
636 |
0.35 |
| chr3_154332843_154334222 | 0.27 |
AI606473 |
expressed sequence AI606473 |
1535 |
0.39 |
| chr10_81642066_81642286 | 0.27 |
Ankrd24 |
ankyrin repeat domain 24 |
1232 |
0.25 |
| chr2_30399608_30399851 | 0.27 |
Dolpp1 |
dolichyl pyrophosphate phosphatase 1 |
2308 |
0.15 |
| chr6_6878154_6880927 | 0.27 |
Dlx5 |
distal-less homeobox 5 |
2528 |
0.19 |
| chr6_30169273_30169444 | 0.27 |
Mir96 |
microRNA 96 |
193 |
0.69 |
| chr1_128804042_128804193 | 0.27 |
Gm26686 |
predicted gene, 26686 |
29696 |
0.19 |
| chr7_45685544_45685874 | 0.27 |
Ntn5 |
netrin 5 |
1280 |
0.18 |
| chr17_35926318_35926661 | 0.27 |
Ppp1r10 |
protein phosphatase 1, regulatory subunit 10 |
5 |
0.92 |
| chr7_45076239_45076453 | 0.27 |
Nosip |
nitric oxide synthase interacting protein |
62 |
0.9 |
| chr7_45075409_45075560 | 0.27 |
Nosip |
nitric oxide synthase interacting protein |
227 |
0.76 |
| chr17_85679503_85681370 | 0.27 |
Six2 |
sine oculis-related homeobox 2 |
7818 |
0.18 |
| chr6_90809120_90810568 | 0.27 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
279 |
0.89 |
| chr6_72273431_72273622 | 0.26 |
Sftpb |
surfactant associated protein B |
31084 |
0.11 |
| chr4_154945283_154945859 | 0.26 |
Hes5 |
hes family bHLH transcription factor 5 |
15352 |
0.1 |
| chr4_13744037_13744679 | 0.26 |
Runx1t1 |
RUNX1 translocation partner 1 |
922 |
0.71 |
| chr17_85072509_85072660 | 0.26 |
Prepl |
prolyl endopeptidase-like |
4890 |
0.21 |
| chr7_46011295_46011446 | 0.26 |
Abcc6 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
10301 |
0.11 |
| chr6_6855562_6856705 | 0.26 |
Gm44094 |
predicted gene, 44094 |
118 |
0.94 |
| chr9_65140865_65142286 | 0.26 |
Igdcc3 |
immunoglobulin superfamily, DCC subclass, member 3 |
363 |
0.83 |
| chr4_48586042_48586294 | 0.26 |
Tmeff1 |
transmembrane protein with EGF-like and two follistatin-like domains 1 |
617 |
0.75 |
| chr2_26314203_26314732 | 0.26 |
Gpsm1 |
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
1048 |
0.36 |
| chr7_35214427_35215048 | 0.26 |
Lrp3 |
low density lipoprotein receptor-related protein 3 |
555 |
0.64 |
| chr2_180506623_180506842 | 0.25 |
Ntsr1 |
neurotensin receptor 1 |
6505 |
0.17 |
| chr1_78189316_78190249 | 0.25 |
Pax3 |
paired box 3 |
7056 |
0.24 |
| chr4_136649051_136649935 | 0.25 |
Gm37583 |
predicted gene, 37583 |
9911 |
0.12 |
| chr1_183141874_183142088 | 0.25 |
Disp1 |
dispatched RND transporter family member 1 |
5480 |
0.24 |
| chr1_132291328_132291479 | 0.25 |
Gm7241 |
predicted pseudogene 7241 |
6950 |
0.12 |
| chr5_34326506_34326657 | 0.25 |
Gm43458 |
predicted gene 43458 |
949 |
0.43 |
| chr10_80192077_80192401 | 0.25 |
Efna2 |
ephrin A2 |
12757 |
0.08 |
| chr7_34653043_34653418 | 0.25 |
Gm12781 |
predicted gene 12781 |
46 |
0.53 |
| chr5_136881600_136883223 | 0.25 |
Col26a1 |
collagen, type XXVI, alpha 1 |
332 |
0.82 |
| chr2_26634327_26634478 | 0.25 |
Dipk1b |
divergent protein kinase domain 1B |
199 |
0.87 |
| chr18_54978491_54979564 | 0.25 |
Zfp608 |
zinc finger protein 608 |
11139 |
0.2 |
| chr10_81324031_81324182 | 0.25 |
Cactin |
cactin, spliceosome C complex subunit |
604 |
0.42 |
| chr11_84725103_84725307 | 0.25 |
1700109G15Rik |
RIKEN cDNA 1700109G15 gene |
4773 |
0.23 |
| chr7_24607911_24608062 | 0.25 |
Phldb3 |
pleckstrin homology like domain, family B, member 3 |
2777 |
0.12 |
| chr3_116421092_116421666 | 0.25 |
Cdc14a |
CDC14 cell division cycle 14A |
1270 |
0.39 |
| chr8_119792153_119792331 | 0.25 |
Meak7 |
MTOR associated protein , eak-7 homolog |
13826 |
0.16 |
| chr11_102264393_102264544 | 0.25 |
Hrob |
homologous recombination factor with OB-fold |
2504 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 0.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.3 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 0.4 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.4 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.4 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.2 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 0.2 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.1 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.1 | 0.3 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.1 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.1 | 0.2 | GO:1901256 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.1 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.6 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.1 | 0.2 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.2 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.1 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.2 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 | 0.2 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.2 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.1 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.2 | GO:0060592 | mammary gland formation(GO:0060592) |
| 0.0 | 0.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.4 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.4 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.1 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.4 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0098700 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.0 | GO:0072197 | ureter morphogenesis(GO:0072197) |
| 0.0 | 0.0 | GO:0021590 | cerebellum maturation(GO:0021590) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.1 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.0 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.4 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.1 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.0 | GO:0072240 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.8 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.0 | 0.0 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.0 | 0.1 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.0 | GO:0010535 | positive regulation of activation of JAK2 kinase activity(GO:0010535) |
| 0.0 | 0.0 | GO:2000054 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:1902661 | regulation of glucose mediated signaling pathway(GO:1902659) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.3 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.0 | GO:0061626 | pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.0 | 0.0 | GO:1990123 | L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.2 | GO:0061430 | bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.0 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.2 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.5 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.1 | GO:1904948 | midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.2 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0003097 | renal water transport(GO:0003097) |
| 0.0 | 0.0 | GO:0010182 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.0 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.0 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.0 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.0 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0097152 | mesenchymal cell apoptotic process(GO:0097152) regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.0 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.4 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.0 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.0 | GO:0060460 | left lung development(GO:0060459) left lung morphogenesis(GO:0060460) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.1 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 0.0 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.0 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.0 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.0 | GO:1903818 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.0 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0033129 | positive regulation of histone phosphorylation(GO:0033129) |
| 0.0 | 0.0 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.0 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.0 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.6 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.0 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.0 | GO:0061197 | fungiform papilla morphogenesis(GO:0061197) |
| 0.0 | 0.0 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.8 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:1990393 | 3M complex(GO:1990393) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.3 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.2 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0015254 | glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.0 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0052759 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.0 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.0 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.0 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.0 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |