Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
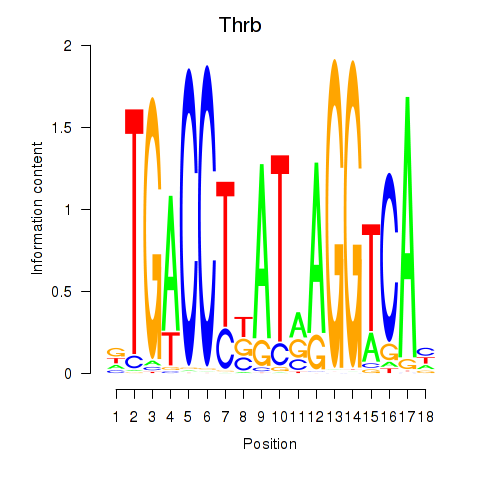
Results for Thrb
Z-value: 0.62
Transcription factors associated with Thrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thrb
|
ENSMUSG00000021779.10 | thyroid hormone receptor beta |
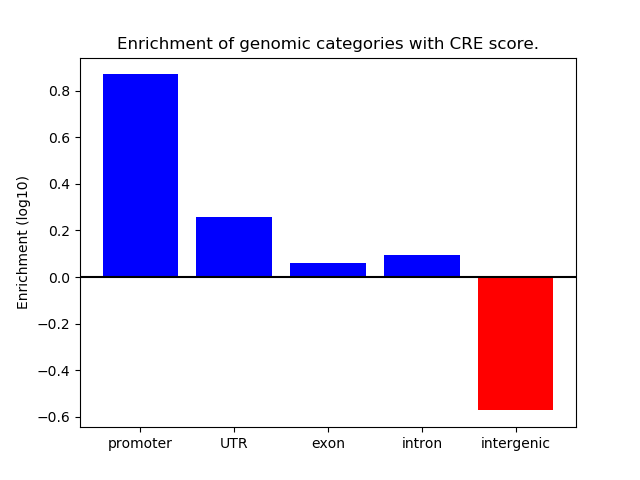
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_17659232_17659747 | Thrb | 772 | 0.780111 | 0.32 | 1.3e-02 | Click! |
| chr14_17632749_17633318 | Thrb | 27228 | 0.255791 | 0.24 | 6.9e-02 | Click! |
| chr14_17660912_17661470 | Thrb | 295 | 0.947354 | 0.21 | 1.1e-01 | Click! |
| chr14_17660241_17660826 | Thrb | 1 | 0.988506 | 0.20 | 1.2e-01 | Click! |
| chr14_17479068_17479220 | Thrb | 181117 | 0.031897 | 0.09 | 4.9e-01 | Click! |
Activity of the Thrb motif across conditions
Conditions sorted by the z-value of the Thrb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
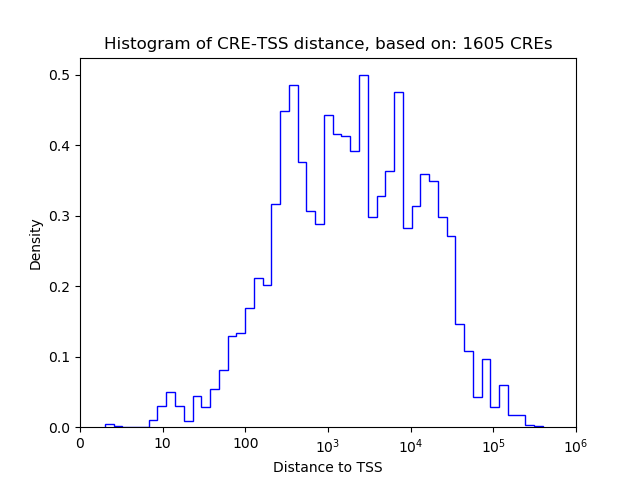
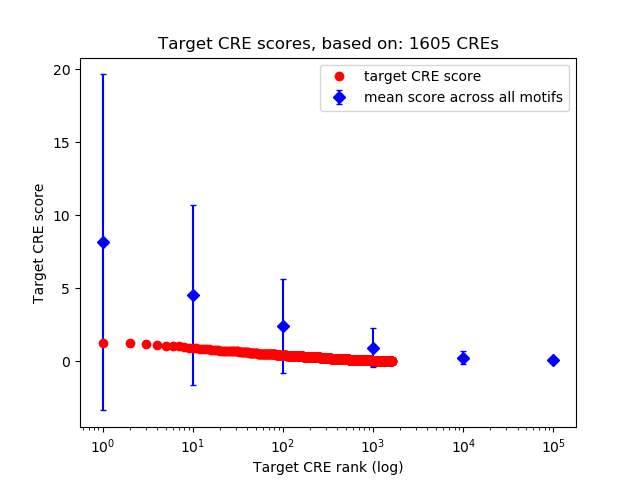
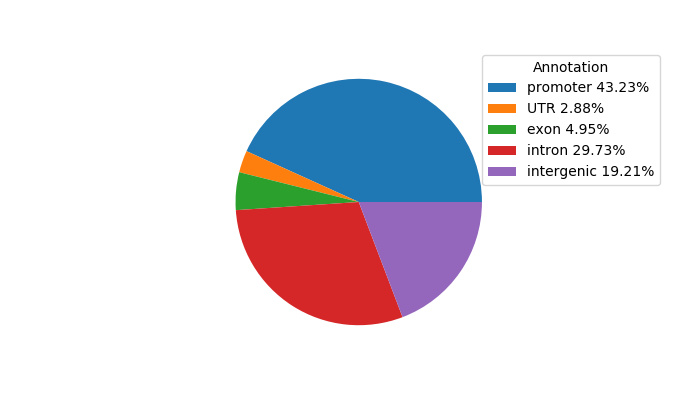
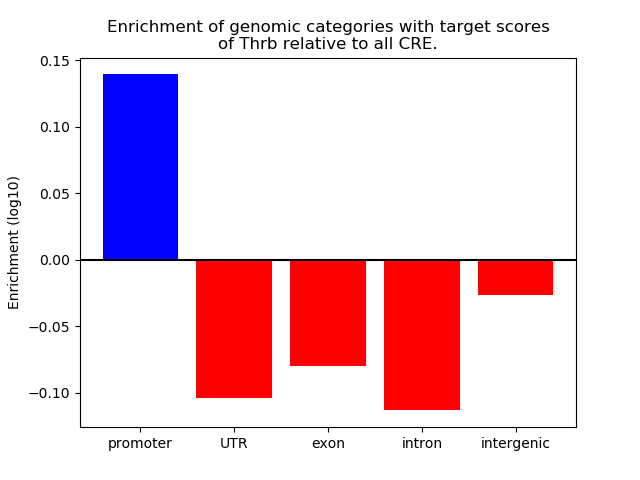
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_98245804_98245958 | 1.25 |
Gm42821 |
predicted gene 42821 |
2718 |
0.21 |
| chr13_49610100_49610769 | 1.25 |
Ogn |
osteoglycin |
2388 |
0.21 |
| chr1_155146107_155146796 | 1.18 |
Mr1 |
major histocompatibility complex, class I-related |
332 |
0.85 |
| chr10_56468695_56469687 | 1.14 |
Gm9118 |
predicted gene 9118 |
28150 |
0.16 |
| chr19_47319497_47320713 | 1.06 |
Sh3pxd2a |
SH3 and PX domains 2A |
5354 |
0.2 |
| chr9_110380536_110381006 | 1.04 |
Scap |
SREBF chaperone |
2478 |
0.17 |
| chr15_27786588_27788615 | 1.02 |
Trio |
triple functional domain (PTPRF interacting) |
1037 |
0.6 |
| chr11_3809703_3809854 | 0.99 |
Gm24013 |
predicted gene, 24013 |
20886 |
0.13 |
| chr9_64017614_64019400 | 0.93 |
Smad6 |
SMAD family member 6 |
1520 |
0.33 |
| chr11_90030594_90032253 | 0.93 |
Tmem100 |
transmembrane protein 100 |
1075 |
0.58 |
| chr16_34923616_34923830 | 0.90 |
Mylk |
myosin, light polypeptide kinase |
6619 |
0.2 |
| chr16_4439334_4439485 | 0.85 |
Adcy9 |
adenylate cyclase 9 |
18911 |
0.19 |
| chr3_122104159_122104612 | 0.83 |
Abca4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
15655 |
0.16 |
| chr15_101128362_101129824 | 0.83 |
Acvrl1 |
activin A receptor, type II-like 1 |
169 |
0.91 |
| chr1_166126774_166128045 | 0.82 |
Dusp27 |
dual specificity phosphatase 27 (putative) |
487 |
0.76 |
| chr7_100288260_100289308 | 0.81 |
P4ha3 |
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide III |
3242 |
0.18 |
| chr12_86000637_86000811 | 0.80 |
Gm26531 |
predicted gene, 26531 |
21703 |
0.16 |
| chr6_71261026_71261496 | 0.77 |
Smyd1 |
SET and MYND domain containing 1 |
971 |
0.38 |
| chr19_38042662_38044080 | 0.74 |
Myof |
myoferlin |
13 |
0.97 |
| chr9_118608620_118608968 | 0.74 |
Itga9 |
integrin alpha 9 |
2104 |
0.28 |
| chr10_56379554_56379705 | 0.72 |
Gja1 |
gap junction protein, alpha 1 |
541 |
0.79 |
| chr2_163396823_163398215 | 0.72 |
Jph2 |
junctophilin 2 |
430 |
0.77 |
| chr11_94498570_94500249 | 0.72 |
Epn3 |
epsin 3 |
289 |
0.85 |
| chr15_76194294_76195998 | 0.71 |
Plec |
plectin |
564 |
0.56 |
| chr15_100401682_100401947 | 0.71 |
Slc11a2 |
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
1429 |
0.28 |
| chr6_99095299_99096526 | 0.71 |
Foxp1 |
forkhead box P1 |
293 |
0.93 |
| chr3_107985653_107986442 | 0.70 |
Gstm2 |
glutathione S-transferase, mu 2 |
313 |
0.73 |
| chr12_76071969_76072878 | 0.70 |
Syne2 |
spectrin repeat containing, nuclear envelope 2 |
407 |
0.88 |
| chr1_71421789_71421940 | 0.68 |
Abca12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
6954 |
0.28 |
| chr16_4521657_4521808 | 0.68 |
Srl |
sarcalumenin |
1331 |
0.38 |
| chr17_49562101_49562252 | 0.68 |
Daam2 |
dishevelled associated activator of morphogenesis 2 |
2130 |
0.39 |
| chr19_43387895_43388797 | 0.68 |
Hpse2 |
heparanase 2 |
9 |
0.98 |
| chr2_118112796_118113528 | 0.67 |
Thbs1 |
thrombospondin 1 |
1286 |
0.33 |
| chr5_124951228_124951428 | 0.66 |
Rflna |
refilin A |
51893 |
0.11 |
| chr3_51378444_51378901 | 0.66 |
Gm5103 |
predicted gene 5103 |
596 |
0.57 |
| chr1_185092269_185092420 | 0.66 |
Gm37662 |
predicted gene, 37662 |
353 |
0.85 |
| chr1_45501357_45501527 | 0.65 |
Col5a2 |
collagen, type V, alpha 2 |
1840 |
0.29 |
| chr19_57417994_57418728 | 0.64 |
Gm50271 |
predicted gene, 50271 |
5133 |
0.19 |
| chr2_164442047_164443132 | 0.64 |
Sdc4 |
syndecan 4 |
597 |
0.53 |
| chr2_60672831_60673749 | 0.61 |
Itgb6 |
integrin beta 6 |
403 |
0.88 |
| chr2_127990589_127990740 | 0.60 |
Acoxl |
acyl-Coenzyme A oxidase-like |
81606 |
0.08 |
| chr7_112155826_112156308 | 0.59 |
Dkk3 |
dickkopf WNT signaling pathway inhibitor 3 |
2990 |
0.34 |
| chr15_79516155_79517393 | 0.58 |
Kdelr3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
363 |
0.76 |
| chr10_107930315_107930872 | 0.58 |
Gm29685 |
predicted gene, 29685 |
16052 |
0.22 |
| chr17_79287021_79288072 | 0.57 |
Gm5230 |
predicted gene 5230 |
29497 |
0.19 |
| chr19_34551724_34552072 | 0.57 |
Ifit2 |
interferon-induced protein with tetratricopeptide repeats 2 |
1188 |
0.33 |
| chr16_43701967_43702307 | 0.56 |
Gm49735 |
predicted gene, 49735 |
8695 |
0.16 |
| chrX_142826741_142826976 | 0.55 |
Tmem164 |
transmembrane protein 164 |
1117 |
0.58 |
| chr3_95673620_95674691 | 0.55 |
Adamtsl4 |
ADAMTS-like 4 |
2996 |
0.14 |
| chr1_28856567_28856718 | 0.55 |
Gm5697 |
predicted gene 5697 |
53921 |
0.15 |
| chr5_135486772_135486923 | 0.55 |
Hip1 |
huntingtin interacting protein 1 |
2411 |
0.23 |
| chr6_17305946_17307996 | 0.54 |
Cav1 |
caveolin 1, caveolae protein |
77 |
0.97 |
| chr15_102102931_102104191 | 0.53 |
Tns2 |
tensin 2 |
573 |
0.62 |
| chr10_34485029_34485207 | 0.53 |
Frk |
fyn-related kinase |
1586 |
0.46 |
| chr7_130258773_130259779 | 0.53 |
Fgfr2 |
fibroblast growth factor receptor 2 |
2581 |
0.38 |
| chr15_76205535_76206787 | 0.53 |
Plec |
plectin |
160 |
0.89 |
| chr3_104565801_104565952 | 0.52 |
Gm26091 |
predicted gene, 26091 |
35363 |
0.09 |
| chr16_29560429_29560777 | 0.52 |
Atp13a4 |
ATPase type 13A4 |
15739 |
0.2 |
| chr11_86586191_86587628 | 0.52 |
Vmp1 |
vacuole membrane protein 1 |
28 |
0.97 |
| chr14_101532225_101532447 | 0.52 |
Tbc1d4 |
TBC1 domain family, member 4 |
25010 |
0.19 |
| chr3_27904926_27905081 | 0.52 |
Tmem212 |
transmembrane protein 212 |
8635 |
0.22 |
| chr2_33975957_33976172 | 0.51 |
Gm13404 |
predicted gene 13404 |
2182 |
0.35 |
| chr13_12098517_12098668 | 0.50 |
Ryr2 |
ryanodine receptor 2, cardiac |
7867 |
0.2 |
| chr4_139460214_139460365 | 0.50 |
Ubr4 |
ubiquitin protein ligase E3 component n-recognin 4 |
2970 |
0.25 |
| chr2_33219879_33220609 | 0.50 |
Angptl2 |
angiopoietin-like 2 |
4127 |
0.19 |
| chr9_110875577_110875744 | 0.50 |
Tmie |
transmembrane inner ear |
361 |
0.73 |
| chr15_77343031_77343868 | 0.50 |
Gm49436 |
predicted gene, 49436 |
23250 |
0.11 |
| chr11_102989753_102990660 | 0.50 |
Dcakd |
dephospho-CoA kinase domain containing |
7342 |
0.12 |
| chr18_11058575_11059292 | 0.50 |
Gata6 |
GATA binding protein 6 |
114 |
0.97 |
| chr18_12270079_12270307 | 0.50 |
Ankrd29 |
ankyrin repeat domain 29 |
1504 |
0.35 |
| chr9_44485094_44485975 | 0.50 |
Bcl9l |
B cell CLL/lymphoma 9-like |
1713 |
0.13 |
| chr18_80360572_80360723 | 0.49 |
D330025C20Rik |
RIKEN cDNA D330025C20 gene |
2134 |
0.23 |
| chr4_147969208_147969498 | 0.49 |
Nppb |
natriuretic peptide type B |
16435 |
0.09 |
| chr5_119836883_119837654 | 0.48 |
Tbx5 |
T-box 5 |
1113 |
0.47 |
| chr6_115990945_115992684 | 0.48 |
Plxnd1 |
plexin D1 |
3191 |
0.2 |
| chr13_31627499_31628464 | 0.48 |
Gm27516 |
predicted gene, 27516 |
1438 |
0.26 |
| chr7_49528137_49528903 | 0.48 |
Nav2 |
neuron navigator 2 |
19672 |
0.23 |
| chr17_56162523_56163227 | 0.47 |
Tnfaip8l1 |
tumor necrosis factor, alpha-induced protein 8-like 1 |
398 |
0.71 |
| chrX_99459091_99459649 | 0.47 |
Gm14808 |
predicted gene 14808 |
345 |
0.87 |
| chr14_26041922_26042457 | 0.47 |
Gm9780 |
predicted gene 9780 |
485 |
0.69 |
| chr17_47178960_47179764 | 0.47 |
Trerf1 |
transcriptional regulating factor 1 |
33025 |
0.17 |
| chr3_87951379_87952516 | 0.46 |
Crabp2 |
cellular retinoic acid binding protein II |
197 |
0.87 |
| chr15_91971921_91972104 | 0.46 |
Gm18979 |
predicted gene, 18979 |
11272 |
0.22 |
| chr17_28345051_28346589 | 0.46 |
Tead3 |
TEA domain family member 3 |
4526 |
0.11 |
| chr9_123480120_123480887 | 0.46 |
Limd1 |
LIM domains containing 1 |
107 |
0.97 |
| chr10_77685958_77687018 | 0.46 |
Tspear |
thrombospondin type laminin G domain and EAR repeats |
81 |
0.9 |
| chr10_81195163_81195734 | 0.46 |
Atcayos |
ataxia, cerebellar, Cayman type, opposite strand |
757 |
0.35 |
| chr14_25902144_25902650 | 0.45 |
Plac9a |
placenta specific 9a |
77 |
0.96 |
| chr4_140580764_140581885 | 0.45 |
Arhgef10l |
Rho guanine nucleotide exchange factor (GEF) 10-like |
478 |
0.83 |
| chr8_124775682_124775942 | 0.45 |
Gm45856 |
predicted gene 45856 |
5921 |
0.12 |
| chr14_54975812_54976152 | 0.44 |
Mir208b |
microRNA 208b |
206 |
0.75 |
| chr16_38361999_38362192 | 0.44 |
Popdc2 |
popeye domain containing 2 |
114 |
0.94 |
| chr5_92901808_92902019 | 0.44 |
Shroom3 |
shroom family member 3 |
3920 |
0.27 |
| chr19_56356629_56356975 | 0.44 |
Gm17197 |
predicted gene 17197 |
7460 |
0.18 |
| chr15_38197898_38198049 | 0.44 |
Gm35167 |
predicted gene, 35167 |
4244 |
0.16 |
| chr10_18197014_18197461 | 0.44 |
Ect2l |
epithelial cell transforming sequence 2 oncogene-like |
4116 |
0.21 |
| chr14_118506460_118506954 | 0.44 |
Abcc4 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
23258 |
0.17 |
| chr5_66497840_66498108 | 0.43 |
Apbb2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
5802 |
0.17 |
| chr4_130014339_130014490 | 0.43 |
Adgrb2 |
adhesion G protein-coupled receptor B2 |
778 |
0.56 |
| chr7_45102549_45104045 | 0.43 |
Fcgrt |
Fc receptor, IgG, alpha chain transporter |
211 |
0.75 |
| chr8_26384770_26384921 | 0.43 |
Gm31898 |
predicted gene, 31898 |
50868 |
0.1 |
| chr16_31878792_31879568 | 0.43 |
Meltf |
melanotransferrin |
370 |
0.79 |
| chr7_25802106_25803222 | 0.42 |
Gm45226 |
predicted gene 45226 |
7511 |
0.08 |
| chr7_130060387_130060669 | 0.41 |
Gm23847 |
predicted gene, 23847 |
25434 |
0.23 |
| chr14_26181467_26182596 | 0.41 |
Plac9b |
placenta specific 9b |
175 |
0.92 |
| chr11_95259399_95259633 | 0.41 |
Tac4 |
tachykinin 4 |
2013 |
0.24 |
| chr4_109550484_109550898 | 0.40 |
Gm22497 |
predicted gene, 22497 |
13652 |
0.15 |
| chr12_86075117_86075268 | 0.40 |
Gm48008 |
predicted gene, 48008 |
3452 |
0.17 |
| chrX_160502969_160503207 | 0.40 |
Phka2 |
phosphorylase kinase alpha 2 |
623 |
0.7 |
| chr2_33714996_33715766 | 0.40 |
9430024E24Rik |
RIKEN cDNA 9430024E24 gene |
3518 |
0.24 |
| chr18_66344941_66345611 | 0.39 |
Ccbe1 |
collagen and calcium binding EGF domains 1 |
42535 |
0.1 |
| chr6_86792679_86793848 | 0.39 |
Anxa4 |
annexin A4 |
301 |
0.87 |
| chr10_26774495_26774696 | 0.39 |
Arhgap18 |
Rho GTPase activating protein 18 |
2048 |
0.32 |
| chr12_56533030_56535358 | 0.39 |
Nkx2-1 |
NK2 homeobox 1 |
912 |
0.36 |
| chr11_33145103_33146186 | 0.39 |
Fgf18 |
fibroblast growth factor 18 |
1151 |
0.43 |
| chr9_62420746_62421699 | 0.39 |
Coro2b |
coronin, actin binding protein, 2B |
6779 |
0.24 |
| chr19_44719535_44720602 | 0.39 |
Gm35610 |
predicted gene, 35610 |
19811 |
0.13 |
| chr4_31823845_31825033 | 0.38 |
Map3k7 |
mitogen-activated protein kinase kinase kinase 7 |
139658 |
0.05 |
| chr13_55830047_55831334 | 0.38 |
Pitx1 |
paired-like homeodomain transcription factor 1 |
735 |
0.57 |
| chr14_26449969_26450211 | 0.38 |
Slmap |
sarcolemma associated protein |
7200 |
0.16 |
| chr12_84875703_84876939 | 0.38 |
Ltbp2 |
latent transforming growth factor beta binding protein 2 |
158 |
0.82 |
| chr5_66069344_66070452 | 0.38 |
Gm43775 |
predicted gene 43775 |
2997 |
0.17 |
| chr16_85150827_85151226 | 0.38 |
Gm49226 |
predicted gene, 49226 |
2197 |
0.3 |
| chr9_108228044_108228195 | 0.38 |
Dag1 |
dystroglycan 1 |
35590 |
0.07 |
| chr3_67373789_67374948 | 0.38 |
Mlf1 |
myeloid leukemia factor 1 |
220 |
0.85 |
| chr17_68165450_68165981 | 0.38 |
Gm49944 |
predicted gene, 49944 |
7841 |
0.22 |
| chr9_95693924_95694752 | 0.37 |
Pcolce2 |
procollagen C-endopeptidase enhancer 2 |
14032 |
0.13 |
| chr10_103027884_103029519 | 0.37 |
Alx1 |
ALX homeobox 1 |
74 |
0.97 |
| chr17_44137199_44137398 | 0.37 |
Clic5 |
chloride intracellular channel 5 |
2530 |
0.33 |
| chr1_152304992_152305188 | 0.37 |
Gm38238 |
predicted gene, 38238 |
12562 |
0.24 |
| chr8_84200619_84201949 | 0.37 |
Gm37352 |
predicted gene, 37352 |
415 |
0.58 |
| chr11_68390483_68390938 | 0.37 |
Ntn1 |
netrin 1 |
3698 |
0.27 |
| chr12_111920312_111920463 | 0.37 |
Gm3565 |
predicted gene 3565 |
10650 |
0.11 |
| chr4_138756785_138756962 | 0.37 |
Pla2g2f |
phospholipase A2, group IIF |
753 |
0.56 |
| chr15_74733696_74733847 | 0.37 |
Lypd2 |
Ly6/Plaur domain containing 2 |
546 |
0.52 |
| chr9_106371285_106372888 | 0.37 |
Dusp7 |
dual specificity phosphatase 7 |
1342 |
0.31 |
| chr5_67669613_67669824 | 0.37 |
Atp8a1 |
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
9684 |
0.14 |
| chr15_41830961_41831808 | 0.36 |
Oxr1 |
oxidation resistance 1 |
391 |
0.87 |
| chr10_66919897_66921448 | 0.36 |
Gm26576 |
predicted gene, 26576 |
370 |
0.51 |
| chr2_38196447_38197128 | 0.36 |
Gm44455 |
predicted gene, 44455 |
28335 |
0.16 |
| chr4_32936919_32937425 | 0.36 |
Ankrd6 |
ankyrin repeat domain 6 |
13654 |
0.15 |
| chr13_31810556_31811961 | 0.36 |
Foxc1 |
forkhead box C1 |
4625 |
0.19 |
| chr12_73052052_73054521 | 0.36 |
Six1 |
sine oculis-related homeobox 1 |
601 |
0.76 |
| chr13_95719581_95720106 | 0.36 |
Gm50469 |
predicted gene, 50469 |
12349 |
0.14 |
| chr3_101835437_101836744 | 0.36 |
Mab21l3 |
mab-21-like 3 |
133 |
0.97 |
| chr6_136660962_136662291 | 0.36 |
Plbd1 |
phospholipase B domain containing 1 |
247 |
0.91 |
| chr3_138069501_138070764 | 0.36 |
1110002E22Rik |
RIKEN cDNA 1110002E22 gene |
1465 |
0.3 |
| chr8_123186072_123187147 | 0.36 |
Dpep1 |
dipeptidase 1 |
367 |
0.69 |
| chr5_36701464_36701615 | 0.35 |
D5Ertd579e |
DNA segment, Chr 5, ERATO Doi 579, expressed |
5515 |
0.15 |
| chr1_186703655_186704733 | 0.35 |
Tgfb2 |
transforming growth factor, beta 2 |
139 |
0.93 |
| chr11_5971864_5972423 | 0.35 |
Camk2b |
calcium/calmodulin-dependent protein kinase II, beta |
11995 |
0.13 |
| chr8_87233933_87234270 | 0.35 |
Gm27169 |
predicted gene 27169 |
31075 |
0.16 |
| chr2_170879045_170879196 | 0.35 |
Gm14263 |
predicted gene 14263 |
75704 |
0.11 |
| chr4_137489689_137489944 | 0.35 |
Hspg2 |
perlecan (heparan sulfate proteoglycan 2) |
21013 |
0.13 |
| chr11_85667588_85668438 | 0.34 |
Bcas3 |
breast carcinoma amplified sequence 3 |
27309 |
0.17 |
| chr3_152943448_152943599 | 0.34 |
Gm16231 |
predicted gene 16231 |
210 |
0.93 |
| chr5_14028529_14028806 | 0.34 |
Gm43519 |
predicted gene 43519 |
773 |
0.63 |
| chr2_30604757_30605755 | 0.34 |
Cstad |
CSA-conditional, T cell activation-dependent protein |
39 |
0.97 |
| chr10_122963294_122964406 | 0.34 |
Mirlet7i |
microRNA let7i |
21874 |
0.16 |
| chr18_75382116_75382565 | 0.33 |
Smad7 |
SMAD family member 7 |
7426 |
0.22 |
| chr7_69740270_69740768 | 0.33 |
Gm7627 |
predicted gene 7627 |
133679 |
0.05 |
| chrX_73929866_73931201 | 0.33 |
Renbp |
renin binding protein |
208 |
0.88 |
| chr10_84530624_84531496 | 0.33 |
Ckap4 |
cytoskeleton-associated protein 4 |
2976 |
0.16 |
| chr11_59223016_59223179 | 0.33 |
Arf1 |
ADP-ribosylation factor 1 |
5065 |
0.11 |
| chr11_87443002_87443199 | 0.33 |
Rnu3b1 |
U3B small nuclear RNA 1 |
137 |
0.89 |
| chr17_26848242_26848532 | 0.33 |
Nkx2-5 |
NK2 homeobox 5 |
3378 |
0.14 |
| chr8_31120852_31121424 | 0.33 |
Rnf122 |
ring finger protein 122 |
9274 |
0.15 |
| chr4_5327298_5327481 | 0.33 |
Gm11782 |
predicted gene 11782 |
223834 |
0.02 |
| chr6_72597498_72597649 | 0.33 |
Elmod3 |
ELMO/CED-12 domain containing 3 |
769 |
0.34 |
| chr17_85691735_85693137 | 0.32 |
CJ186046Rik |
Riken cDNA CJ186046 gene |
1193 |
0.46 |
| chr7_16823026_16823360 | 0.32 |
Strn4 |
striatin, calmodulin binding protein 4 |
122 |
0.93 |
| chr1_15791844_15792059 | 0.32 |
Terf1 |
telomeric repeat binding factor 1 |
13695 |
0.16 |
| chr5_145115072_145115244 | 0.32 |
Arpc1b |
actin related protein 2/3 complex, subunit 1B |
842 |
0.42 |
| chrX_161997301_161997452 | 0.32 |
Gm26317 |
predicted gene, 26317 |
72081 |
0.11 |
| chr10_111801109_111801260 | 0.32 |
Gm47865 |
predicted gene, 47865 |
1331 |
0.33 |
| chr5_67427677_67428127 | 0.32 |
Gm42632 |
predicted gene 42632 |
91 |
0.42 |
| chr1_82586379_82587667 | 0.32 |
Col4a3 |
collagen, type IV, alpha 3 |
47 |
0.67 |
| chr8_57799694_57800553 | 0.32 |
Galntl6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
3533 |
0.27 |
| chr2_71626794_71626945 | 0.32 |
Dlx2 |
distal-less homeobox 2 |
80115 |
0.06 |
| chrX_12886648_12887120 | 0.31 |
Gm25063 |
predicted gene, 25063 |
16522 |
0.17 |
| chr13_55018577_55018728 | 0.31 |
Hk3 |
hexokinase 3 |
2252 |
0.24 |
| chr9_84503048_84503199 | 0.31 |
Gm39388 |
predicted gene, 39388 |
74837 |
0.1 |
| chr5_24366542_24366693 | 0.31 |
Nos3 |
nitric oxide synthase 3, endothelial cell |
1807 |
0.18 |
| chr15_74997196_74997485 | 0.31 |
Ly6a |
lymphocyte antigen 6 complex, locus A |
294 |
0.78 |
| chr1_162076839_162077220 | 0.31 |
Gm37767 |
predicted gene, 37767 |
11439 |
0.19 |
| chr7_101377838_101379825 | 0.31 |
Arap1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
462 |
0.71 |
| chr9_53342768_53342919 | 0.30 |
Exph5 |
exophilin 5 |
1654 |
0.36 |
| chr12_103336858_103338251 | 0.30 |
Gm15523 |
predicted gene 15523 |
648 |
0.43 |
| chr2_38514387_38514655 | 0.30 |
Nek6 |
NIMA (never in mitosis gene a)-related expressed kinase 6 |
60 |
0.96 |
| chr14_21903607_21905272 | 0.30 |
4931407E12Rik |
RIKEN cDNA 4931407E12 gene |
14673 |
0.14 |
| chr17_33778675_33780469 | 0.30 |
Angptl4 |
angiopoietin-like 4 |
429 |
0.65 |
| chr17_13683677_13683962 | 0.30 |
Gm16046 |
predicted gene 16046 |
303 |
0.87 |
| chr10_12547923_12549201 | 0.30 |
Utrn |
utrophin |
317 |
0.94 |
| chr1_97802358_97802693 | 0.30 |
Pam |
peptidylglycine alpha-amidating monooxygenase |
23460 |
0.16 |
| chr9_50751156_50752468 | 0.30 |
Cryab |
crystallin, alpha B |
65 |
0.86 |
| chr5_143000025_143001100 | 0.30 |
Rnf216 |
ring finger protein 216 |
16212 |
0.14 |
| chr2_174288589_174289023 | 0.30 |
Gnasas1 |
GNAS antisense RNA 1 |
3371 |
0.16 |
| chr4_76369301_76369461 | 0.30 |
Gm11252 |
predicted gene 11252 |
23773 |
0.21 |
| chr10_81195811_81196102 | 0.30 |
Atcayos |
ataxia, cerebellar, Cayman type, opposite strand |
1265 |
0.19 |
| chr19_5842574_5845856 | 0.30 |
Neat1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
1044 |
0.25 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.2 | 0.7 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 0.6 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.2 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.6 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.2 | 0.5 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.2 | 0.5 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.4 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.6 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 0.8 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.3 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 0.3 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.7 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 0.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.3 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.3 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 0.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.1 | 0.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.5 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.3 | GO:0035983 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.2 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.1 | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.2 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.2 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.2 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.4 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.2 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.1 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 1.0 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.5 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.2 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.3 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.1 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.1 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.0 | 0.2 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.0 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.0 | 0.2 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0000087 | mitotic M phase(GO:0000087) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.0 | 0.3 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.4 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.0 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 | 0.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.2 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.0 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.0 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.0 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.0 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.0 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.0 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0071404 | cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.0 | 0.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.0 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.0 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 | 0.3 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.2 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.0 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.0 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.0 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.1 | GO:1990845 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.0 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.0 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.1 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.0 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.4 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.2 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 0.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.2 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.1 | 0.2 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0052724 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 1.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.5 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.0 | GO:0030351 | inositol-1,3,4,5,6-pentakisphosphate 3-phosphatase activity(GO:0030351) inositol-1,4,5,6-tetrakisphosphate 6-phosphatase activity(GO:0030352) inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.0 | 0.1 | GO:0052622 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 1.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.0 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.4 | GO:0051723 | protein methylesterase activity(GO:0051723) |
| 0.0 | 1.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.0 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.0 | GO:0019198 | transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.5 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.0 | REACTOME METABOLISM OF RNA | Genes involved in Metabolism of RNA |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |