Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
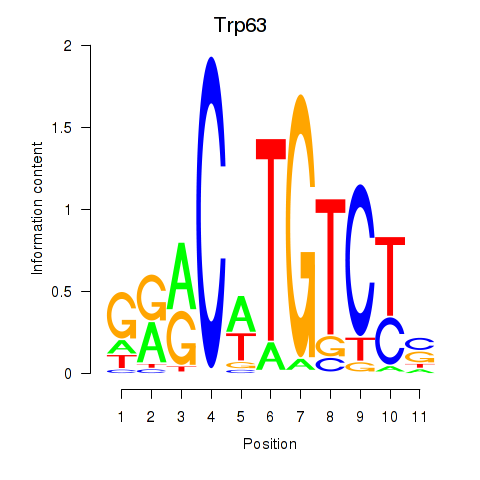
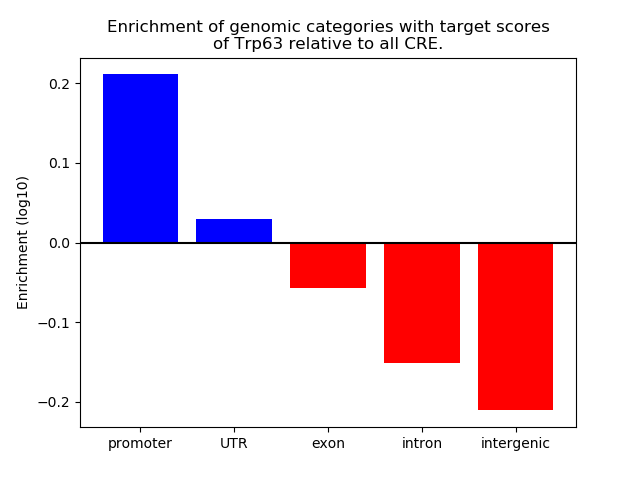
Results for Trp63
Z-value: 0.29
Transcription factors associated with Trp63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp63
|
ENSMUSG00000022510.8 | transformation related protein 63 |
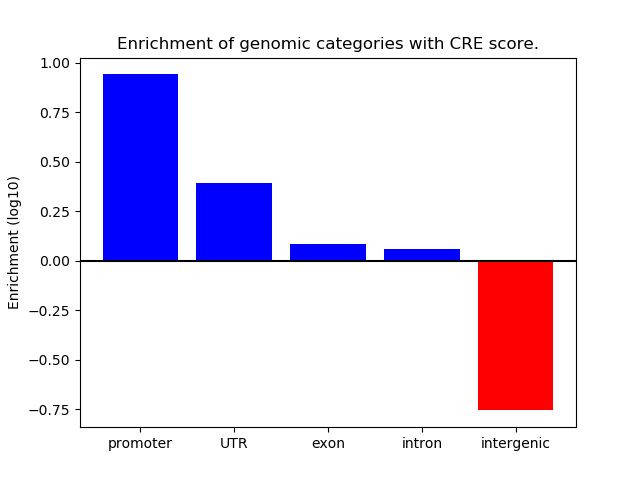
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_25684291_25684442 | Trp63 | 601 | 0.776410 | 0.38 | 2.6e-03 | Click! |
| chr16_25683728_25684269 | Trp63 | 233 | 0.942184 | 0.26 | 4.8e-02 | Click! |
| chr16_25804319_25804643 | Trp63 | 2565 | 0.377228 | 0.09 | 5.1e-01 | Click! |
| chr16_25806162_25806335 | Trp63 | 4332 | 0.308764 | -0.08 | 5.3e-01 | Click! |
| chr16_25804012_25804163 | Trp63 | 2171 | 0.412444 | 0.07 | 6.1e-01 | Click! |
Activity of the Trp63 motif across conditions
Conditions sorted by the z-value of the Trp63 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_90738322_90739000 | 0.26 |
Mrap |
melanocortin 2 receptor accessory protein |
337 |
0.85 |
| chr2_32081622_32082932 | 0.26 |
Fam78a |
family with sequence similarity 78, member A |
1506 |
0.26 |
| chr15_98601965_98602724 | 0.24 |
Adcy6 |
adenylate cyclase 6 |
2387 |
0.14 |
| chr12_74013636_74013876 | 0.24 |
Syt16 |
synaptotagmin XVI |
15957 |
0.17 |
| chr15_77754933_77755315 | 0.21 |
Apol8 |
apolipoprotein L 8 |
115 |
0.93 |
| chr6_117918083_117918316 | 0.19 |
Gm43863 |
predicted gene, 43863 |
477 |
0.57 |
| chr15_83169184_83169335 | 0.19 |
Cyb5r3 |
cytochrome b5 reductase 3 |
918 |
0.4 |
| chr11_98910013_98910427 | 0.19 |
Cdc6 |
cell division cycle 6 |
2069 |
0.19 |
| chr16_19982574_19983084 | 0.18 |
Klhl6 |
kelch-like 6 |
101 |
0.97 |
| chr2_91120847_91121416 | 0.18 |
Mybpc3 |
myosin binding protein C, cardiac |
2987 |
0.16 |
| chr14_31576784_31577279 | 0.18 |
Colq |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
352 |
0.85 |
| chr8_122282435_122282616 | 0.17 |
Zfpm1 |
zinc finger protein, multitype 1 |
384 |
0.83 |
| chr8_80497324_80498362 | 0.17 |
Gypa |
glycophorin A |
4062 |
0.27 |
| chr8_70476235_70476894 | 0.17 |
Klhl26 |
kelch-like 26 |
341 |
0.75 |
| chr1_135839302_135839453 | 0.17 |
Tnnt2 |
troponin T2, cardiac |
1322 |
0.34 |
| chr11_32288387_32289175 | 0.16 |
Hbq1b |
hemoglobin, theta 1B |
1780 |
0.2 |
| chr10_128494842_128495139 | 0.16 |
Myl6b |
myosin, light polypeptide 6B |
566 |
0.4 |
| chr17_47595220_47595679 | 0.16 |
Ccnd3 |
cyclin D3 |
619 |
0.57 |
| chr3_152477886_152478987 | 0.16 |
Ak5 |
adenylate kinase 5 |
79 |
0.97 |
| chr8_57513444_57513905 | 0.15 |
Hmgb2 |
high mobility group box 2 |
1126 |
0.33 |
| chr7_44468072_44469106 | 0.15 |
Josd2 |
Josephin domain containing 2 |
237 |
0.75 |
| chr4_141421681_141422558 | 0.15 |
Hspb7 |
heat shock protein family, member 7 (cardiovascular) |
1340 |
0.25 |
| chr15_36606762_36607275 | 0.15 |
Pabpc1 |
poly(A) binding protein, cytoplasmic 1 |
1552 |
0.29 |
| chr11_55184284_55185056 | 0.15 |
Slc36a2 |
solute carrier family 36 (proton/amino acid symporter), member 2 |
407 |
0.78 |
| chr6_47191483_47191919 | 0.14 |
Cntnap2 |
contactin associated protein-like 2 |
52686 |
0.16 |
| chr1_51288641_51290950 | 0.14 |
Cavin2 |
caveolae associated 2 |
669 |
0.72 |
| chr2_59829432_59829769 | 0.14 |
Tanc1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
12140 |
0.22 |
| chr11_114198650_114199013 | 0.14 |
1700092K14Rik |
RIKEN cDNA 1700092K14 gene |
573 |
0.74 |
| chr9_103305951_103306102 | 0.14 |
Topbp1 |
topoisomerase (DNA) II binding protein 1 |
523 |
0.61 |
| chr1_135837715_135838209 | 0.14 |
Tnnt2 |
troponin T2, cardiac |
1549 |
0.29 |
| chr11_117784224_117784441 | 0.14 |
Gm11723 |
predicted gene 11723 |
723 |
0.4 |
| chr12_28804949_28805160 | 0.14 |
Gm48905 |
predicted gene, 48905 |
5084 |
0.18 |
| chr17_24250421_24251368 | 0.14 |
Ccnf |
cyclin F |
424 |
0.68 |
| chr9_75441517_75442270 | 0.13 |
Leo1 |
Leo1, Paf1/RNA polymerase II complex component |
357 |
0.81 |
| chr6_146220976_146221548 | 0.13 |
Itpr2 |
inositol 1,4,5-triphosphate receptor 2 |
6281 |
0.26 |
| chr3_153689877_153690164 | 0.13 |
Gm22206 |
predicted gene, 22206 |
18891 |
0.16 |
| chr7_17027108_17027849 | 0.13 |
Ppp5c |
protein phosphatase 5, catalytic subunit |
137 |
0.93 |
| chr19_55926597_55927105 | 0.12 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
28542 |
0.21 |
| chr1_82288297_82288775 | 0.12 |
Irs1 |
insulin receptor substrate 1 |
2880 |
0.26 |
| chr19_43520527_43520933 | 0.12 |
Got1 |
glutamic-oxaloacetic transaminase 1, soluble |
3755 |
0.15 |
| chr9_21526198_21526743 | 0.12 |
AB124611 |
cDNA sequence AB124611 |
242 |
0.85 |
| chr2_26012349_26012671 | 0.12 |
Ubac1 |
ubiquitin associated domain containing 1 |
1557 |
0.29 |
| chr11_90726819_90728183 | 0.12 |
Tom1l1 |
target of myb1-like 1 (chicken) |
39135 |
0.15 |
| chr7_24373502_24374343 | 0.12 |
Kcnn4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
3584 |
0.11 |
| chr3_138067510_138069146 | 0.12 |
Gm5105 |
predicted gene 5105 |
940 |
0.46 |
| chrX_11664777_11665621 | 0.12 |
Gm14513 |
predicted gene 14513 |
18925 |
0.24 |
| chr3_101551232_101552184 | 0.12 |
Atp1a1 |
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
25852 |
0.16 |
| chr12_24963377_24963763 | 0.12 |
Kidins220 |
kinase D-interacting substrate 220 |
11355 |
0.18 |
| chr11_69947014_69948171 | 0.12 |
Slc2a4 |
solute carrier family 2 (facilitated glucose transporter), member 4 |
382 |
0.6 |
| chr19_40376409_40376771 | 0.12 |
Sorbs1 |
sorbin and SH3 domain containing 1 |
284 |
0.93 |
| chr6_31612888_31614126 | 0.11 |
Gm43154 |
predicted gene 43154 |
8218 |
0.19 |
| chr6_125349094_125349245 | 0.11 |
Tnfrsf1a |
tumor necrosis factor receptor superfamily, member 1a |
193 |
0.91 |
| chr13_75711286_75711461 | 0.11 |
Ell2 |
elongation factor RNA polymerase II 2 |
3662 |
0.15 |
| chr19_10578600_10578808 | 0.11 |
Cyb561a3 |
cytochrome b561 family, member A3 |
692 |
0.43 |
| chr9_21015148_21016366 | 0.11 |
Icam1 |
intercellular adhesion molecule 1 |
228 |
0.81 |
| chr12_112942194_112942430 | 0.11 |
Nudt14 |
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
188 |
0.88 |
| chr11_32250575_32250853 | 0.11 |
Nprl3 |
nitrogen permease regulator-like 3 |
436 |
0.71 |
| chr4_88550254_88550926 | 0.11 |
Ifna15 |
interferon alpha 15 |
7655 |
0.1 |
| chr15_77815874_77816114 | 0.11 |
Myh9 |
myosin, heavy polypeptide 9, non-muscle |
2763 |
0.21 |
| chr16_37539321_37539726 | 0.11 |
Gtf2e1 |
general transcription factor II E, polypeptide 1 (alpha subunit) |
253 |
0.6 |
| chr13_28770503_28770945 | 0.11 |
Gm17528 |
predicted gene, 17528 |
56399 |
0.12 |
| chr18_35847749_35850271 | 0.11 |
Cxxc5 |
CXXC finger 5 |
5677 |
0.11 |
| chr6_137410715_137411297 | 0.11 |
Ptpro |
protein tyrosine phosphatase, receptor type, O |
247 |
0.94 |
| chr1_135840170_135841177 | 0.11 |
Tnnt2 |
troponin T2, cardiac |
26 |
0.97 |
| chr7_136894579_136895285 | 0.11 |
Mgmt |
O-6-methylguanine-DNA methyltransferase |
318 |
0.92 |
| chr9_113930385_113930581 | 0.11 |
Ubp1 |
upstream binding protein 1 |
451 |
0.84 |
| chr7_4866688_4866875 | 0.11 |
Isoc2b |
isochorismatase domain containing 2b |
588 |
0.52 |
| chr11_55411877_55412028 | 0.11 |
Sparc |
secreted acidic cysteine rich glycoprotein |
7946 |
0.16 |
| chr1_193382120_193382498 | 0.11 |
Camk1g |
calcium/calmodulin-dependent protein kinase I gamma |
12011 |
0.15 |
| chr13_29854543_29854861 | 0.10 |
Cdkal1 |
CDK5 regulatory subunit associated protein 1-like 1 |
729 |
0.77 |
| chr8_45658542_45659771 | 0.10 |
Sorbs2 |
sorbin and SH3 domain containing 2 |
352 |
0.89 |
| chr11_58918885_58919318 | 0.10 |
Btnl10 |
butyrophilin-like 10 |
1044 |
0.25 |
| chr15_83147790_83147941 | 0.10 |
Poldip3 |
polymerase (DNA-directed), delta interacting protein 3 |
1436 |
0.23 |
| chr3_69005043_69005730 | 0.10 |
Smc4 |
structural maintenance of chromosomes 4 |
371 |
0.66 |
| chr6_141273994_141274145 | 0.10 |
Gm28523 |
predicted gene 28523 |
24060 |
0.19 |
| chr3_100921543_100921984 | 0.10 |
Trim45 |
tripartite motif-containing 45 |
439 |
0.82 |
| chr9_56505872_56506042 | 0.10 |
Gm47175 |
predicted gene, 47175 |
704 |
0.65 |
| chr8_122719956_122721464 | 0.10 |
C230057M02Rik |
RIKEN cDNA C230057M02 gene |
17801 |
0.09 |
| chr13_111894572_111895253 | 0.10 |
Gm9025 |
predicted gene 9025 |
10735 |
0.15 |
| chr8_85432612_85432832 | 0.10 |
1700051O22Rik |
RIKEN cDNA 1700051O22 Gene |
36 |
0.56 |
| chr1_78568355_78568575 | 0.10 |
Mogat1 |
monoacylglycerol O-acyltransferase 1 |
30837 |
0.13 |
| chr17_12764355_12764670 | 0.10 |
Igf2r |
insulin-like growth factor 2 receptor |
5152 |
0.13 |
| chr6_38900943_38901392 | 0.10 |
Tbxas1 |
thromboxane A synthase 1, platelet |
17813 |
0.19 |
| chr8_75089284_75089957 | 0.10 |
Hmox1 |
heme oxygenase 1 |
4001 |
0.16 |
| chr6_120467285_120467457 | 0.10 |
Il17ra |
interleukin 17 receptor A |
4124 |
0.17 |
| chr11_32671444_32671658 | 0.10 |
Fbxw11 |
F-box and WD-40 domain protein 11 |
28647 |
0.18 |
| chr15_85634869_85635356 | 0.10 |
Gm49539 |
predicted gene, 49539 |
10971 |
0.15 |
| chr7_110777775_110778052 | 0.10 |
Ampd3 |
adenosine monophosphate deaminase 3 |
223 |
0.91 |
| chr7_19226915_19227555 | 0.10 |
Opa3 |
optic atrophy 3 |
1099 |
0.27 |
| chr2_119972478_119973221 | 0.10 |
Mapkbp1 |
mitogen-activated protein kinase binding protein 1 |
54 |
0.96 |
| chr14_63946096_63946930 | 0.10 |
Sox7 |
SRY (sex determining region Y)-box 7 |
2840 |
0.24 |
| chr1_138912105_138912653 | 0.10 |
Gm3933 |
predicted gene 3933 |
3664 |
0.15 |
| chrX_132102482_132102633 | 0.10 |
Gm7058 |
predicted gene 7058 |
67550 |
0.13 |
| chr5_33272179_33273173 | 0.10 |
Ctbp1 |
C-terminal binding protein 1 |
1903 |
0.28 |
| chr11_119392783_119393622 | 0.09 |
Rnf213 |
ring finger protein 213 |
102 |
0.95 |
| chr14_65832957_65833923 | 0.09 |
Esco2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
468 |
0.79 |
| chr10_127289828_127290445 | 0.09 |
Gm4189 |
predicted gene 4189 |
588 |
0.31 |
| chr3_139308377_139308557 | 0.09 |
Stpg2 |
sperm tail PG rich repeat containing 2 |
102574 |
0.07 |
| chr12_84709717_84709962 | 0.09 |
Syndig1l |
synapse differentiation inducing 1 like |
11008 |
0.15 |
| chr7_92873732_92874248 | 0.09 |
Ddias |
DNA damage-induced apoptosis suppressor |
186 |
0.77 |
| chr16_18428270_18428753 | 0.09 |
Txnrd2 |
thioredoxin reductase 2 |
186 |
0.88 |
| chr16_3846654_3847068 | 0.09 |
Zfp174 |
zinc finger protein 174 |
407 |
0.71 |
| chr4_107253592_107253822 | 0.09 |
Hspb11 |
heat shock protein family B (small), member 11 |
114 |
0.58 |
| chr3_131334397_131335246 | 0.09 |
Sgms2 |
sphingomyelin synthase 2 |
10112 |
0.15 |
| chr12_109454496_109454673 | 0.09 |
Dlk1 |
delta like non-canonical Notch ligand 1 |
389 |
0.74 |
| chr3_84189087_84189700 | 0.09 |
Trim2 |
tripartite motif-containing 2 |
1130 |
0.52 |
| chr3_159847798_159848044 | 0.09 |
Wls |
wntless WNT ligand secretion mediator |
530 |
0.83 |
| chr11_8501968_8502413 | 0.09 |
Tns3 |
tensin 3 |
33515 |
0.23 |
| chr11_45926491_45926959 | 0.09 |
Lsm11 |
U7 snRNP-specific Sm-like protein LSM11 |
18210 |
0.14 |
| chr6_119196453_119196655 | 0.09 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
136 |
0.96 |
| chr5_116420481_116421179 | 0.09 |
Hspb8 |
heat shock protein 8 |
2034 |
0.22 |
| chr5_135777784_135778296 | 0.09 |
Styxl1 |
serine/threonine/tyrosine interacting-like 1 |
198 |
0.71 |
| chr6_55203433_55204260 | 0.09 |
Mindy4 |
MINDY lysine 48 deubiquitinase 4 |
463 |
0.81 |
| chr8_94986231_94987228 | 0.09 |
Adgrg1 |
adhesion G protein-coupled receptor G1 |
1161 |
0.36 |
| chr4_135353535_135353968 | 0.09 |
Srrm1 |
serine/arginine repetitive matrix 1 |
430 |
0.73 |
| chr7_102246806_102247734 | 0.09 |
Rhog |
ras homolog family member G |
2818 |
0.17 |
| chr1_166124763_166124914 | 0.09 |
Dusp27 |
dual specificity phosphatase 27 (putative) |
3058 |
0.21 |
| chr16_91169710_91170388 | 0.09 |
Gm49613 |
predicted gene, 49613 |
5285 |
0.13 |
| chr1_74049307_74049652 | 0.09 |
Tns1 |
tensin 1 |
12322 |
0.21 |
| chr10_127290769_127291873 | 0.09 |
Ddit3 |
DNA-damage inducible transcript 3 |
492 |
0.37 |
| chr1_135768530_135768972 | 0.09 |
Phlda3 |
pleckstrin homology like domain, family A, member 3 |
1617 |
0.31 |
| chr4_115092064_115093406 | 0.09 |
Pdzk1ip1 |
PDZK1 interacting protein 1 |
1803 |
0.28 |
| chr12_51058382_51058886 | 0.09 |
Gm22088 |
predicted gene, 22088 |
27053 |
0.18 |
| chr11_101094859_101095385 | 0.09 |
Psmc3ip |
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
254 |
0.81 |
| chr19_43921380_43921848 | 0.09 |
Gm50217 |
predicted gene, 50217 |
1141 |
0.4 |
| chr11_101627997_101628632 | 0.08 |
Rdm1 |
RAD52 motif 1 |
75 |
0.9 |
| chr1_58029648_58030887 | 0.08 |
Aox1 |
aldehyde oxidase 1 |
303 |
0.89 |
| chr5_117133717_117134877 | 0.08 |
Taok3 |
TAO kinase 3 |
656 |
0.62 |
| chr14_54252799_54253017 | 0.08 |
Dad1 |
defender against cell death 1 |
1006 |
0.28 |
| chr5_75607943_75608525 | 0.08 |
Kit |
KIT proto-oncogene receptor tyrosine kinase |
2340 |
0.34 |
| chr4_132973841_132975368 | 0.08 |
Fgr |
FGR proto-oncogene, Src family tyrosine kinase |
502 |
0.76 |
| chr10_80899333_80899562 | 0.08 |
Timm13 |
translocase of inner mitochondrial membrane 13 |
1335 |
0.24 |
| chr3_41081759_41082908 | 0.08 |
Pgrmc2 |
progesterone receptor membrane component 2 |
713 |
0.68 |
| chr17_33555776_33556414 | 0.08 |
Myo1f |
myosin IF |
330 |
0.85 |
| chr7_48991870_48992129 | 0.08 |
Nav2 |
neuron navigator 2 |
32902 |
0.14 |
| chr16_75906235_75906386 | 0.08 |
Samsn1 |
SAM domain, SH3 domain and nuclear localization signals, 1 |
2969 |
0.33 |
| chr9_44110354_44111126 | 0.08 |
Gm47327 |
predicted gene, 47327 |
169 |
0.8 |
| chr3_137342527_137342893 | 0.08 |
Emcn |
endomucin |
1536 |
0.49 |
| chr7_141442950_141444299 | 0.08 |
Pidd1 |
p53 induced death domain protein 1 |
268 |
0.74 |
| chr2_9877985_9878236 | 0.08 |
Gata3 |
GATA binding protein 3 |
490 |
0.56 |
| chr17_79348163_79348612 | 0.08 |
Cdc42ep3 |
CDC42 effector protein (Rho GTPase binding) 3 |
4620 |
0.24 |
| chr5_115509295_115510185 | 0.08 |
Pxn |
paxillin |
2884 |
0.12 |
| chr1_59579877_59580151 | 0.08 |
Gm973 |
predicted gene 973 |
2382 |
0.23 |
| chr9_123366984_123367730 | 0.08 |
Lars2 |
leucyl-tRNA synthetase, mitochondrial |
354 |
0.88 |
| chr7_141010753_141011069 | 0.08 |
Ifitm3 |
interferon induced transmembrane protein 3 |
141 |
0.89 |
| chr8_120535808_120536639 | 0.08 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
366 |
0.73 |
| chr13_25562934_25563111 | 0.08 |
Gm11349 |
predicted gene 11349 |
20364 |
0.25 |
| chr11_69095229_69096278 | 0.08 |
Per1 |
period circadian clock 1 |
536 |
0.52 |
| chr19_24527047_24527577 | 0.08 |
Pip5k1b |
phosphatidylinositol-4-phosphate 5-kinase, type 1 beta |
28477 |
0.16 |
| chr2_163354062_163354414 | 0.08 |
Tox2 |
TOX high mobility group box family member 2 |
33860 |
0.12 |
| chr1_82288792_82289141 | 0.08 |
Irs1 |
insulin receptor substrate 1 |
2450 |
0.28 |
| chr5_149184724_149185776 | 0.08 |
Uspl1 |
ubiquitin specific peptidase like 1 |
469 |
0.47 |
| chrX_164074168_164074319 | 0.08 |
Siah1b |
siah E3 ubiquitin protein ligase 1B |
1878 |
0.31 |
| chr8_18738571_18739759 | 0.08 |
Angpt2 |
angiopoietin 2 |
2397 |
0.3 |
| chr17_45573465_45573903 | 0.08 |
Hsp90ab1 |
heat shock protein 90 alpha (cytosolic), class B member 1 |
413 |
0.69 |
| chr11_100967422_100967790 | 0.08 |
Cavin1 |
caveolae associated 1 |
2945 |
0.18 |
| chr7_144489028_144489179 | 0.08 |
Ppfia1 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
234 |
0.92 |
| chr13_37482944_37483624 | 0.08 |
Gm47732 |
predicted gene, 47732 |
899 |
0.41 |
| chr6_125491631_125492511 | 0.07 |
Gm26728 |
predicted gene, 26728 |
2353 |
0.22 |
| chr7_46554047_46554198 | 0.07 |
Gm15700 |
predicted gene 15700 |
7179 |
0.15 |
| chr2_120154741_120155150 | 0.07 |
Ehd4 |
EH-domain containing 4 |
339 |
0.87 |
| chr11_11828454_11829202 | 0.07 |
Ddc |
dopa decarboxylase |
5907 |
0.2 |
| chr12_110022984_110023602 | 0.07 |
Gm34667 |
predicted gene, 34667 |
580 |
0.67 |
| chr11_103158442_103159191 | 0.07 |
Fmnl1 |
formin-like 1 |
12291 |
0.11 |
| chr16_91466275_91466861 | 0.07 |
Gm49626 |
predicted gene, 49626 |
1443 |
0.19 |
| chr1_125678382_125678847 | 0.07 |
Gpr39 |
G protein-coupled receptor 39 |
1619 |
0.44 |
| chr10_8427639_8427823 | 0.07 |
Ust |
uronyl-2-sulfotransferase |
91094 |
0.09 |
| chr10_60752050_60752803 | 0.07 |
Slc29a3 |
solute carrier family 29 (nucleoside transporters), member 3 |
356 |
0.89 |
| chr13_43158853_43159330 | 0.07 |
Tbc1d7 |
TBC1 domain family, member 7 |
217 |
0.94 |
| chr10_91083048_91083580 | 0.07 |
Ikbip |
IKBKB interacting protein |
194 |
0.79 |
| chr9_104063750_104064058 | 0.07 |
Acad11 |
acyl-Coenzyme A dehydrogenase family, member 11 |
171 |
0.84 |
| chr2_52619696_52619936 | 0.07 |
Bloc1s2-ps |
biogenesis of lysosomal organelles complex-1, subunit 2, pseudogene |
62 |
0.98 |
| chr13_3866217_3866406 | 0.07 |
Calm5 |
calmodulin 5 |
12043 |
0.11 |
| chr14_70521525_70521739 | 0.07 |
Sftpc |
surfactant associated protein C |
544 |
0.57 |
| chr13_20900415_20900566 | 0.07 |
Gm25605 |
predicted gene, 25605 |
55736 |
0.12 |
| chr16_93885679_93885894 | 0.07 |
Chaf1b |
chromatin assembly factor 1, subunit B (p60) |
1584 |
0.28 |
| chr2_103895883_103896333 | 0.07 |
Gm13876 |
predicted gene 13876 |
7784 |
0.12 |
| chr15_73791907_73792230 | 0.07 |
Ndufb4c |
NADH:ubiquinone oxidoreductase subunit B4C |
17828 |
0.15 |
| chr10_71347309_71347708 | 0.07 |
Ipmk |
inositol polyphosphate multikinase |
255 |
0.88 |
| chr3_98267489_98267783 | 0.07 |
Hmgcs2 |
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 |
12799 |
0.13 |
| chr3_96404203_96405307 | 0.07 |
Gm26654 |
predicted gene, 26654 |
2374 |
0.09 |
| chr11_89056720_89056879 | 0.07 |
Dgke |
diacylglycerol kinase, epsilon |
3949 |
0.15 |
| chr2_91964507_91965860 | 0.07 |
Dgkz |
diacylglycerol kinase zeta |
404 |
0.78 |
| chr19_24378594_24378946 | 0.07 |
Pip5k1bos |
phosphatidylinositol-4-phosphate 5-kinase, type 1 beta, opposite strand |
37239 |
0.15 |
| chr11_68850735_68851370 | 0.07 |
Ndel1 |
nudE neurodevelopment protein 1 like 1 |
1989 |
0.24 |
| chr14_7774273_7774663 | 0.07 |
Gm10044 |
predicted gene 10044 |
329 |
0.85 |
| chr17_81064219_81065026 | 0.07 |
Thumpd2 |
THUMP domain containing 2 |
445 |
0.89 |
| chr9_51009335_51009739 | 0.07 |
Sik2 |
salt inducible kinase 2 |
464 |
0.81 |
| chr2_167833410_167833763 | 0.07 |
1200007C13Rik |
RIKEN cDNA 1200007C13 gene |
60 |
0.97 |
| chr3_99965564_99965715 | 0.07 |
Rpl36-ps7 |
ribosomal protein L36, pseudogene 7 |
42721 |
0.16 |
| chr2_30981192_30981821 | 0.07 |
BC005624 |
cDNA sequence BC005624 |
435 |
0.61 |
| chr17_33018251_33018434 | 0.07 |
Zfp763 |
zinc finger protein 763 |
14826 |
0.12 |
| chr6_54329484_54329647 | 0.07 |
9130019P16Rik |
RIKEN cDNA 9130019P16 gene |
2224 |
0.26 |
| chr12_73908042_73908546 | 0.07 |
Hif1a |
hypoxia inducible factor 1, alpha subunit |
390 |
0.84 |
| chr1_179581898_179582278 | 0.07 |
Cnst |
consortin, connexin sorting protein |
11302 |
0.17 |
| chr11_69408850_69409533 | 0.07 |
Kdm6b |
KDM1 lysine (K)-specific demethylase 6B |
4484 |
0.09 |
| chr2_145933841_145934674 | 0.07 |
Crnkl1 |
crooked neck pre-mRNA splicing factor 1 |
457 |
0.5 |
| chr7_118853827_118853990 | 0.07 |
Knop1 |
lysine rich nucleolar protein 1 |
1705 |
0.25 |
| chr19_57358602_57359019 | 0.06 |
Fam160b1 |
family with sequence similarity 160, member B1 |
1870 |
0.25 |
| chr2_163658129_163658339 | 0.06 |
Pkig |
protein kinase inhibitor, gamma |
152 |
0.94 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.2 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0060374 | mast cell differentiation(GO:0060374) |
| 0.0 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.1 | GO:0061356 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) |
| 0.0 | 0.1 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.1 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.0 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.0 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.0 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.0 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.0 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0030313 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0052758 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |