Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
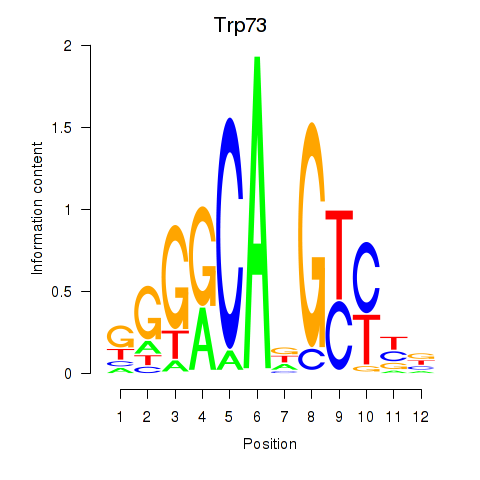
Results for Trp73
Z-value: 0.58
Transcription factors associated with Trp73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Trp73
|
ENSMUSG00000029026.10 | transformation related protein 73 |
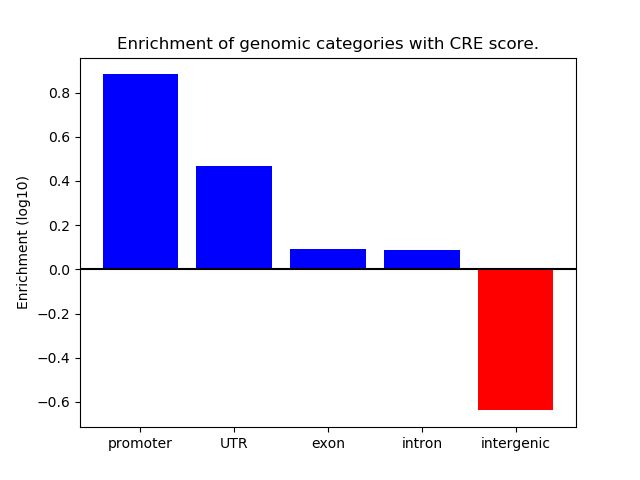
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_154121987_154122318 | Trp73 | 5547 | 0.123269 | 0.28 | 2.8e-02 | Click! |
| chr4_154079845_154079996 | Trp73 | 5402 | 0.119575 | 0.21 | 1.0e-01 | Click! |
| chr4_154080712_154082009 | Trp73 | 3962 | 0.132498 | 0.21 | 1.0e-01 | Click! |
| chr4_154109522_154109673 | Trp73 | 7008 | 0.118316 | -0.21 | 1.1e-01 | Click! |
| chr4_154080558_154080712 | Trp73 | 4687 | 0.124682 | 0.17 | 2.1e-01 | Click! |
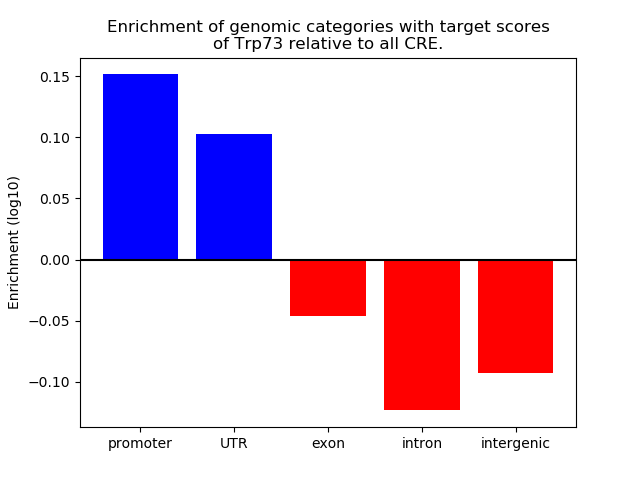
Activity of the Trp73 motif across conditions
Conditions sorted by the z-value of the Trp73 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
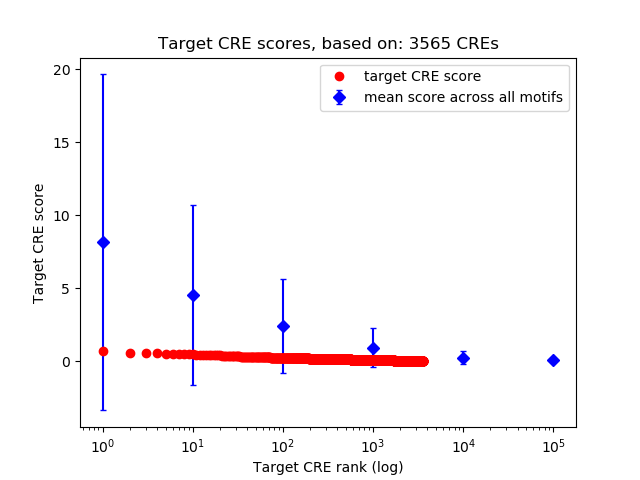
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_170109295_170110836 | 0.71 |
Ddr2 |
discoidin domain receptor family, member 2 |
436 |
0.82 |
| chr17_84183089_84183918 | 0.60 |
Gm36279 |
predicted gene, 36279 |
2253 |
0.25 |
| chr4_58550341_58551468 | 0.57 |
Lpar1 |
lysophosphatidic acid receptor 1 |
2280 |
0.31 |
| chr17_24721463_24721641 | 0.55 |
Rps2 |
ribosomal protein S2 |
103 |
0.81 |
| chr9_21015148_21016366 | 0.52 |
Icam1 |
intercellular adhesion molecule 1 |
228 |
0.81 |
| chr7_122160942_122161093 | 0.52 |
Plk1 |
polo like kinase 1 |
1436 |
0.21 |
| chr7_141505665_141505980 | 0.51 |
Gm45416 |
predicted gene 45416 |
12781 |
0.07 |
| chr18_76034867_76035898 | 0.50 |
1700003O11Rik |
RIKEN cDNA 1700003O11 gene |
19705 |
0.19 |
| chr7_30314557_30315674 | 0.48 |
Syne4 |
spectrin repeat containing, nuclear envelope family member 4 |
35 |
0.92 |
| chr4_98223398_98223549 | 0.47 |
Gm12692 |
predicted gene 12692 |
11817 |
0.21 |
| chr11_99023355_99024178 | 0.45 |
Top2a |
topoisomerase (DNA) II alpha |
412 |
0.74 |
| chr6_41703225_41703532 | 0.44 |
Kel |
Kell blood group |
961 |
0.44 |
| chr10_69925300_69927130 | 0.43 |
Ank3 |
ankyrin 3, epithelial |
82 |
0.99 |
| chr2_84936571_84938205 | 0.43 |
Slc43a3 |
solute carrier family 43, member 3 |
498 |
0.71 |
| chr2_27037174_27037325 | 0.43 |
Gm13398 |
predicted gene 13398 |
36 |
0.95 |
| chr5_119685576_119687800 | 0.41 |
Tbx3os2 |
T-box 3, opposite strand 2 |
4530 |
0.17 |
| chr18_64484992_64485250 | 0.41 |
Fech |
ferrochelatase |
3820 |
0.19 |
| chr7_19796098_19797425 | 0.40 |
Cblc |
Casitas B-lineage lymphoma c |
32 |
0.93 |
| chr4_119188779_119189142 | 0.40 |
Ermap |
erythroblast membrane-associated protein |
213 |
0.86 |
| chr13_21917785_21919224 | 0.40 |
Gm44456 |
predicted gene, 44456 |
6512 |
0.07 |
| chr15_83169184_83169335 | 0.38 |
Cyb5r3 |
cytochrome b5 reductase 3 |
918 |
0.4 |
| chr10_44526709_44528700 | 0.37 |
Prdm1 |
PR domain containing 1, with ZNF domain |
797 |
0.61 |
| chr11_95914404_95916171 | 0.36 |
B4galnt2 |
beta-1,4-N-acetyl-galactosaminyl transferase 2 |
396 |
0.75 |
| chr8_121082801_121085531 | 0.36 |
Foxf1 |
forkhead box F1 |
220 |
0.71 |
| chr9_58248942_58249216 | 0.35 |
Pml |
promyelocytic leukemia |
544 |
0.66 |
| chr10_120364470_120365452 | 0.35 |
1700006J14Rik |
RIKEN cDNA 1700006J14 gene |
748 |
0.63 |
| chr8_27104092_27104394 | 0.35 |
Adgra2 |
adhesion G protein-coupled receptor A2 |
9890 |
0.11 |
| chr17_46242717_46243000 | 0.35 |
Polr1c |
polymerase (RNA) I polypeptide C |
3715 |
0.1 |
| chr2_25195300_25197035 | 0.34 |
Tor4a |
torsin family 4, member A |
592 |
0.43 |
| chr19_10422292_10423153 | 0.34 |
Gm50268 |
predicted gene, 50268 |
17175 |
0.13 |
| chr3_84467720_84468259 | 0.34 |
Fhdc1 |
FH2 domain containing 1 |
10993 |
0.23 |
| chr15_83088765_83089618 | 0.33 |
Serhl |
serine hydrolase-like |
315 |
0.84 |
| chr5_134867554_134868195 | 0.33 |
Tmem270 |
transmembrane protein 270 |
38859 |
0.08 |
| chr9_44655251_44655818 | 0.33 |
Gm47238 |
predicted gene, 47238 |
4613 |
0.08 |
| chr16_34031173_34031775 | 0.33 |
Kalrn |
kalirin, RhoGEF kinase |
14980 |
0.19 |
| chr8_120535808_120536639 | 0.32 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
366 |
0.73 |
| chr11_87729732_87730395 | 0.32 |
Rnf43 |
ring finger protein 43 |
689 |
0.49 |
| chr9_83834733_83835396 | 0.32 |
Ttk |
Ttk protein kinase |
298 |
0.91 |
| chr17_27558079_27558612 | 0.31 |
Hmga1 |
high mobility group AT-hook 1 |
1650 |
0.16 |
| chr4_155061781_155061932 | 0.31 |
Plch2 |
phospholipase C, eta 2 |
5072 |
0.13 |
| chr7_34798114_34799420 | 0.31 |
Chst8 |
carbohydrate sulfotransferase 8 |
13905 |
0.21 |
| chr5_114969022_114970855 | 0.31 |
Hnf1aos1 |
HNF1 homeobox A, opposite strand 1 |
18 |
0.91 |
| chr12_105038616_105039126 | 0.31 |
Glrx5 |
glutaredoxin 5 |
3655 |
0.12 |
| chr2_34873990_34875403 | 0.31 |
Cutal |
cutA divalent cation tolerance homolog-like |
178 |
0.6 |
| chr8_117335167_117335696 | 0.31 |
Cmip |
c-Maf inducing protein |
13739 |
0.25 |
| chr4_134016138_134016675 | 0.30 |
Gm13061 |
predicted gene 13061 |
1741 |
0.21 |
| chr15_36606762_36607275 | 0.29 |
Pabpc1 |
poly(A) binding protein, cytoplasmic 1 |
1552 |
0.29 |
| chr1_130740150_130740628 | 0.29 |
AA986860 |
expressed sequence AA986860 |
117 |
0.89 |
| chr18_60756688_60756839 | 0.29 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
8113 |
0.14 |
| chr13_15466077_15468087 | 0.29 |
Gli3 |
GLI-Kruppel family member GLI3 |
3102 |
0.22 |
| chr7_78913499_78914279 | 0.28 |
Isg20 |
interferon-stimulated protein |
92 |
0.95 |
| chr2_167671877_167672567 | 0.28 |
Gm14320 |
predicted gene 14320 |
10113 |
0.1 |
| chr3_89731508_89731661 | 0.28 |
Adar |
adenosine deaminase, RNA-specific |
836 |
0.52 |
| chr2_144009074_144009285 | 0.28 |
Rrbp1 |
ribosome binding protein 1 |
2084 |
0.3 |
| chr10_81166672_81166823 | 0.28 |
Pias4 |
protein inhibitor of activated STAT 4 |
967 |
0.28 |
| chr4_118429529_118430133 | 0.28 |
Elovl1 |
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
130 |
0.92 |
| chr2_35610114_35610843 | 0.28 |
Dab2ip |
disabled 2 interacting protein |
11503 |
0.2 |
| chr4_135727528_135728972 | 0.28 |
Il22ra1 |
interleukin 22 receptor, alpha 1 |
78 |
0.96 |
| chr5_118327546_118328619 | 0.27 |
Gm25076 |
predicted gene, 25076 |
61633 |
0.09 |
| chr7_136894579_136895285 | 0.27 |
Mgmt |
O-6-methylguanine-DNA methyltransferase |
318 |
0.92 |
| chr11_115832199_115833263 | 0.27 |
Llgl2 |
LLGL2 scribble cell polarity complex component |
763 |
0.46 |
| chr2_93644408_93646515 | 0.27 |
Alx4 |
aristaless-like homeobox 4 |
3073 |
0.32 |
| chr11_114047821_114048516 | 0.27 |
Sdk2 |
sidekick cell adhesion molecule 2 |
17807 |
0.22 |
| chr5_31191554_31191824 | 0.27 |
Eif2b4 |
eukaryotic translation initiation factor 2B, subunit 4 delta |
42 |
0.92 |
| chr11_115076163_115076314 | 0.27 |
Cd300e |
CD300E molecule |
14061 |
0.1 |
| chr6_72215599_72215943 | 0.27 |
Atoh8 |
atonal bHLH transcription factor 8 |
18766 |
0.15 |
| chr18_64484316_64484544 | 0.27 |
Fech |
ferrochelatase |
4511 |
0.18 |
| chr11_97439854_97442222 | 0.27 |
Arhgap23 |
Rho GTPase activating protein 23 |
4753 |
0.18 |
| chr15_76459155_76459306 | 0.27 |
Scx |
scleraxis |
1710 |
0.17 |
| chr2_93176536_93177325 | 0.26 |
Trp53i11 |
transformation related protein 53 inducible protein 11 |
10618 |
0.22 |
| chr7_48881832_48882357 | 0.26 |
E2f8 |
E2F transcription factor 8 |
498 |
0.56 |
| chr6_4488753_4490330 | 0.26 |
Gm37883 |
predicted gene, 37883 |
5343 |
0.17 |
| chr15_76695245_76695396 | 0.26 |
Gpt |
glutamic pyruvic transaminase, soluble |
396 |
0.63 |
| chr14_54574275_54574961 | 0.26 |
Ajuba |
ajuba LIM protein |
2536 |
0.13 |
| chrX_50842772_50843829 | 0.26 |
Stk26 |
serine/threonine kinase 26 |
1859 |
0.47 |
| chr8_91071785_91071936 | 0.25 |
Rbl2 |
RB transcriptional corepressor like 2 |
1702 |
0.27 |
| chr9_59288905_59289056 | 0.25 |
Adpgk |
ADP-dependent glucokinase |
2578 |
0.3 |
| chr9_20973703_20974045 | 0.25 |
S1pr2 |
sphingosine-1-phosphate receptor 2 |
104 |
0.92 |
| chr17_48271543_48271790 | 0.25 |
Treml4 |
triggering receptor expressed on myeloid cells-like 4 |
773 |
0.51 |
| chr6_23247289_23250418 | 0.25 |
Fezf1 |
Fez family zinc finger 1 |
491 |
0.76 |
| chr5_139352673_139352961 | 0.25 |
Cyp2w1 |
cytochrome P450, family 2, subfamily w, polypeptide 1 |
200 |
0.88 |
| chr10_84457678_84458129 | 0.25 |
Gm47962 |
predicted gene, 47962 |
17077 |
0.12 |
| chrX_112697857_112698775 | 0.25 |
Pof1b |
premature ovarian failure 1B |
335 |
0.89 |
| chr11_6266682_6266833 | 0.25 |
Ddx56 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 56 |
365 |
0.76 |
| chr18_32036159_32036926 | 0.25 |
Myo7b |
myosin VIIB |
419 |
0.76 |
| chr9_35424475_35426234 | 0.25 |
Cdon |
cell adhesion molecule-related/down-regulated by oncogenes |
1766 |
0.31 |
| chr10_81340818_81340969 | 0.24 |
Gipc3 |
GIPC PDZ domain containing family, member 3 |
2373 |
0.09 |
| chr15_78581151_78582050 | 0.24 |
Gm36738 |
predicted gene, 36738 |
16 |
0.96 |
| chr3_154329308_154331090 | 0.24 |
Lhx8 |
LIM homeobox protein 8 |
140 |
0.88 |
| chr9_66988141_66989423 | 0.24 |
Gm24225 |
predicted gene, 24225 |
7597 |
0.16 |
| chr5_64810297_64813272 | 0.24 |
Klf3 |
Kruppel-like factor 3 (basic) |
555 |
0.71 |
| chr13_23298244_23298395 | 0.24 |
Gm11333 |
predicted gene 11333 |
270 |
0.8 |
| chr1_75124216_75124367 | 0.24 |
Nhej1 |
non-homologous end joining factor 1 |
909 |
0.38 |
| chr14_76622687_76623498 | 0.24 |
Fkbp1a-ps1 |
FK506 binding protein 1a, pseudogene 1 |
13772 |
0.2 |
| chr9_48744945_48745425 | 0.24 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
90760 |
0.08 |
| chr2_26928944_26929117 | 0.24 |
Surf4 |
surfeit gene 4 |
4353 |
0.08 |
| chr11_87725925_87727087 | 0.24 |
Rnf43 |
ring finger protein 43 |
920 |
0.38 |
| chr7_46849299_46849599 | 0.24 |
Ldha |
lactate dehydrogenase A |
462 |
0.66 |
| chr11_72960644_72962160 | 0.24 |
Atp2a3 |
ATPase, Ca++ transporting, ubiquitous |
112 |
0.95 |
| chr4_155830034_155830298 | 0.23 |
Aurkaip1 |
aurora kinase A interacting protein 1 |
1106 |
0.23 |
| chr5_142440144_142441204 | 0.23 |
Ap5z1 |
adaptor-related protein complex 5, zeta 1 subunit |
23270 |
0.17 |
| chr7_46036795_46037045 | 0.23 |
Nomo1 |
nodal modulator 1 |
3222 |
0.15 |
| chr6_88842552_88843314 | 0.23 |
Gm15612 |
predicted gene 15612 |
79 |
0.88 |
| chr7_45718994_45719157 | 0.23 |
Rpl18 |
ribosomal protein L18 |
77 |
0.88 |
| chr8_22806092_22806678 | 0.23 |
1700041G16Rik |
RIKEN cDNA 1700041G16 gene |
619 |
0.46 |
| chr8_88432600_88432751 | 0.23 |
Gm45497 |
predicted gene 45497 |
41554 |
0.15 |
| chr7_13034712_13035156 | 0.23 |
Chmp2a |
charged multivesicular body protein 2A |
127 |
0.89 |
| chrX_164074168_164074319 | 0.23 |
Siah1b |
siah E3 ubiquitin protein ligase 1B |
1878 |
0.31 |
| chr17_46073528_46074596 | 0.23 |
Gm36200 |
predicted gene, 36200 |
10204 |
0.13 |
| chr5_88679199_88679350 | 0.23 |
Grsf1 |
G-rich RNA sequence binding factor 1 |
3103 |
0.22 |
| chr18_75434066_75434315 | 0.23 |
Smad7 |
SMAD family member 7 |
59276 |
0.12 |
| chr17_24762097_24762281 | 0.23 |
Hs3st6 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
9173 |
0.07 |
| chr17_35502353_35502632 | 0.23 |
Pou5f1 |
POU domain, class 5, transcription factor 1 |
3526 |
0.09 |
| chr3_129215238_129216443 | 0.23 |
Pitx2 |
paired-like homeodomain transcription factor 2 |
1565 |
0.34 |
| chr10_80160846_80161444 | 0.23 |
Cirbp |
cold inducible RNA binding protein |
4840 |
0.09 |
| chr9_67617390_67617685 | 0.23 |
Tln2 |
talin 2 |
57834 |
0.12 |
| chr7_80210085_80210365 | 0.23 |
Gm45206 |
predicted gene 45206 |
831 |
0.42 |
| chr14_99301852_99302138 | 0.23 |
Klf5 |
Kruppel-like factor 5 |
2841 |
0.23 |
| chr3_137978506_137978972 | 0.22 |
Dapp1 |
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
2791 |
0.18 |
| chr11_18870055_18872175 | 0.22 |
8430419K02Rik |
RIKEN cDNA 8430419K02 gene |
1040 |
0.49 |
| chr2_93648842_93648993 | 0.22 |
Alx4 |
aristaless-like homeobox 4 |
6529 |
0.25 |
| chr11_86630813_86631114 | 0.22 |
Gm11478 |
predicted gene 11478 |
63 |
0.97 |
| chr14_54406578_54406953 | 0.22 |
Slc7a7 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
2695 |
0.13 |
| chr11_76309545_76309696 | 0.22 |
Mrm3 |
mitochondrial rRNA methyltransferase 3 |
60408 |
0.09 |
| chr7_100911201_100912201 | 0.22 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
17011 |
0.13 |
| chr9_62840810_62841073 | 0.22 |
Gm10653 |
predicted gene 10653 |
1406 |
0.29 |
| chr13_49545764_49546368 | 0.22 |
Aspn |
asporin |
1579 |
0.33 |
| chr7_4629257_4630354 | 0.22 |
Tmem86b |
transmembrane protein 86B |
382 |
0.66 |
| chr4_154024404_154026596 | 0.22 |
Smim1 |
small integral membrane protein 1 |
116 |
0.93 |
| chr4_126147745_126147906 | 0.22 |
Eva1b |
eva-1 homolog B (C. elegans) |
81 |
0.94 |
| chr7_126430318_126430469 | 0.22 |
Rabep2 |
rabaptin, RAB GTPase binding effector protein 2 |
953 |
0.36 |
| chr17_35051116_35051817 | 0.22 |
Clic1 |
chloride intracellular channel 1 |
984 |
0.19 |
| chr11_32288387_32289175 | 0.22 |
Hbq1b |
hemoglobin, theta 1B |
1780 |
0.2 |
| chr7_35121962_35122959 | 0.22 |
Gm45091 |
predicted gene 45091 |
2855 |
0.13 |
| chr1_171373446_171373749 | 0.21 |
Nectin4 |
nectin cell adhesion molecule 4 |
3242 |
0.09 |
| chr7_141220739_141221753 | 0.21 |
Mir210 |
microRNA 210 |
247 |
0.79 |
| chr11_70650522_70651860 | 0.21 |
Rnf167 |
ring finger protein 167 |
1255 |
0.18 |
| chr1_39471300_39471739 | 0.21 |
Tbc1d8 |
TBC1 domain family, member 8 |
7142 |
0.16 |
| chr7_123368027_123368481 | 0.21 |
Lcmt1 |
leucine carboxyl methyltransferase 1 |
1530 |
0.31 |
| chr17_44193392_44193678 | 0.21 |
Clic5 |
chloride intracellular channel 5 |
4951 |
0.3 |
| chr3_96322102_96322647 | 0.21 |
Gm24830 |
predicted gene, 24830 |
6051 |
0.06 |
| chr2_103957804_103957955 | 0.21 |
Lmo2 |
LIM domain only 2 |
107 |
0.96 |
| chr7_45003101_45003252 | 0.21 |
Irf3 |
interferon regulatory factor 3 |
1789 |
0.12 |
| chr17_85690573_85690815 | 0.21 |
Six2 |
sine oculis-related homeobox 2 |
2420 |
0.25 |
| chr16_29990679_29990830 | 0.21 |
Gm1968 |
predicted gene 1968 |
11570 |
0.16 |
| chr11_60810014_60810742 | 0.21 |
Shmt1 |
serine hydroxymethyltransferase 1 (soluble) |
713 |
0.45 |
| chr10_81464705_81465157 | 0.21 |
Gm16105 |
predicted gene 16105 |
3772 |
0.08 |
| chr4_126324302_126324453 | 0.21 |
Gm12945 |
predicted gene 12945 |
778 |
0.35 |
| chr10_84885973_84886950 | 0.21 |
Ric8b |
RIC8 guanine nucleotide exchange factor B |
31155 |
0.17 |
| chr11_120231879_120233008 | 0.21 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
504 |
0.52 |
| chr10_79946794_79947379 | 0.21 |
Arid3a |
AT rich interactive domain 3A (BRIGHT-like) |
807 |
0.33 |
| chr10_79776981_79777609 | 0.21 |
Fstl3 |
follistatin-like 3 |
3 |
0.93 |
| chr2_9877985_9878236 | 0.21 |
Gata3 |
GATA binding protein 3 |
490 |
0.56 |
| chr17_28255542_28255762 | 0.21 |
Ppard |
peroxisome proliferator activator receptor delta |
16056 |
0.1 |
| chr6_124717857_124719000 | 0.21 |
Mir200c |
microRNA 200c |
38 |
0.7 |
| chr4_87804582_87805116 | 0.21 |
Mllt3 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
1445 |
0.57 |
| chr1_74410022_74410173 | 0.21 |
Vil1 |
villin 1 |
721 |
0.52 |
| chr14_69271950_69272335 | 0.21 |
Gm20236 |
predicted gene, 20236 |
9998 |
0.09 |
| chr14_69490201_69490585 | 0.21 |
Gm37094 |
predicted gene, 37094 |
9997 |
0.1 |
| chr11_4227013_4227476 | 0.21 |
Gm11956 |
predicted gene 11956 |
4021 |
0.11 |
| chr12_110706254_110707091 | 0.21 |
Hsp90aa1 |
heat shock protein 90, alpha (cytosolic), class A member 1 |
3944 |
0.15 |
| chr8_70438199_70438350 | 0.20 |
Crtc1 |
CREB regulated transcription coactivator 1 |
1301 |
0.3 |
| chr10_82762742_82763996 | 0.20 |
Nfyb |
nuclear transcription factor-Y beta |
277 |
0.86 |
| chr11_69907985_69908217 | 0.20 |
Neurl4 |
neuralized E3 ubiquitin protein ligase 4 |
534 |
0.46 |
| chrX_100774452_100775080 | 0.20 |
Dlg3 |
discs large MAGUK scaffold protein 3 |
25 |
0.97 |
| chr6_125422981_125423227 | 0.20 |
Cd9 |
CD9 antigen |
39920 |
0.1 |
| chr15_102235091_102236313 | 0.20 |
Itgb7 |
integrin beta 7 |
3758 |
0.11 |
| chr7_44853717_44855163 | 0.20 |
Akt1s1 |
AKT1 substrate 1 (proline-rich) |
1869 |
0.12 |
| chr9_57298079_57298333 | 0.20 |
Gm18996 |
predicted gene, 18996 |
31209 |
0.12 |
| chr7_19821864_19823009 | 0.20 |
Bcl3 |
B cell leukemia/lymphoma 3 |
279 |
0.77 |
| chr8_85084307_85084458 | 0.20 |
Man2b1 |
mannosidase 2, alpha B1 |
94 |
0.89 |
| chr1_16228040_16228294 | 0.20 |
Gm28095 |
predicted gene 28095 |
339 |
0.88 |
| chr2_167563437_167563946 | 0.20 |
Gm11476 |
predicted gene 11476 |
10377 |
0.12 |
| chr15_44457385_44458529 | 0.20 |
Pkhd1l1 |
polycystic kidney and hepatic disease 1-like 1 |
404 |
0.83 |
| chr17_46444584_46445824 | 0.20 |
Gm5093 |
predicted gene 5093 |
5107 |
0.11 |
| chr6_117918083_117918316 | 0.20 |
Gm43863 |
predicted gene, 43863 |
477 |
0.57 |
| chr17_47595220_47595679 | 0.20 |
Ccnd3 |
cyclin D3 |
619 |
0.57 |
| chr8_94705293_94705444 | 0.20 |
Pllp |
plasma membrane proteolipid |
9090 |
0.12 |
| chr4_106804374_106805428 | 0.20 |
Acot11 |
acyl-CoA thioesterase 11 |
97 |
0.96 |
| chr3_66980595_66981350 | 0.20 |
Shox2 |
short stature homeobox 2 |
169 |
0.76 |
| chr1_167242473_167242624 | 0.20 |
Gm37982 |
predicted gene, 37982 |
91 |
0.89 |
| chr2_173061138_173061289 | 0.19 |
Gm14453 |
predicted gene 14453 |
26633 |
0.12 |
| chr5_91850572_91850781 | 0.19 |
Gm5558 |
predicted gene 5558 |
43603 |
0.1 |
| chr16_20730381_20730693 | 0.19 |
Thpo |
thrombopoietin |
14 |
0.94 |
| chr2_168839664_168840615 | 0.19 |
Gm25214 |
predicted gene, 25214 |
42029 |
0.13 |
| chr9_102248394_102248672 | 0.19 |
Gm37260 |
predicted gene, 37260 |
25203 |
0.17 |
| chr4_106314250_106314401 | 0.19 |
Usp24 |
ubiquitin specific peptidase 24 |
1888 |
0.3 |
| chr15_97781945_97783108 | 0.19 |
Slc48a1 |
solute carrier family 48 (heme transporter), member 1 |
1829 |
0.24 |
| chr6_72620259_72620562 | 0.19 |
Tgoln1 |
trans-golgi network protein |
3410 |
0.1 |
| chrX_150547515_150548479 | 0.19 |
Alas2 |
aminolevulinic acid synthase 2, erythroid |
538 |
0.44 |
| chr12_85192731_85192882 | 0.19 |
Gm45930 |
predicted gene, 45930 |
5001 |
0.12 |
| chr11_117799058_117799291 | 0.19 |
6030468B19Rik |
RIKEN cDNA 6030468B19 gene |
1514 |
0.19 |
| chr8_27177069_27177905 | 0.19 |
Rab11fip1 |
RAB11 family interacting protein 1 (class I) |
2841 |
0.14 |
| chr7_141442950_141444299 | 0.19 |
Pidd1 |
p53 induced death domain protein 1 |
268 |
0.74 |
| chr16_31878792_31879568 | 0.19 |
Meltf |
melanotransferrin |
370 |
0.79 |
| chr4_57611408_57611803 | 0.19 |
Pakap |
paralemmin A kinase anchor protein |
26218 |
0.23 |
| chr5_106465135_106467300 | 0.19 |
Gm26872 |
predicted gene, 26872 |
4790 |
0.19 |
| chr3_108730269_108730718 | 0.19 |
Gpsm2 |
G-protein signalling modulator 2 (AGS3-like, C. elegans) |
8184 |
0.14 |
| chr3_129194987_129195449 | 0.19 |
Pitx2 |
paired-like homeodomain transcription factor 2 |
4660 |
0.2 |
| chr16_33922248_33922575 | 0.19 |
Itgb5 |
integrin beta 5 |
75 |
0.97 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.5 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 0.3 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.1 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.6 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.3 | GO:0048371 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.1 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.3 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.1 | 0.2 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 | 0.2 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.1 | 0.2 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.1 | 0.3 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.1 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.3 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.1 | 0.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.4 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 | 0.5 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.1 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.2 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.1 | 0.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 0.2 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 0.2 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.5 | GO:0018904 | ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.2 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.2 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.2 | GO:0019660 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.0 | 0.4 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.2 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.2 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.1 | GO:0071335 | hair follicle cell proliferation(GO:0071335) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.1 | GO:1990123 | L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:1903911 | positive regulation of receptor clustering(GO:1903911) regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.1 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0034443 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.4 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.0 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0003180 | aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) |
| 0.0 | 0.1 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.7 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.0 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.1 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.1 | GO:0090237 | regulation of arachidonic acid secretion(GO:0090237) |
| 0.0 | 0.1 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.0 | 0.1 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.3 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.0 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0042520 | positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.2 | GO:1902403 | signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.1 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.1 | GO:0052490 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.0 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.0 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.0 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.0 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.0 | 0.0 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.0 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 | 0.1 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.3 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0033121 | regulation of purine nucleotide catabolic process(GO:0033121) |
| 0.0 | 0.0 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.0 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.0 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.1 | GO:0000820 | regulation of glutamine family amino acid metabolic process(GO:0000820) |
| 0.0 | 0.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.0 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.0 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.0 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.0 | GO:1900086 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.0 | GO:0031052 | programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.0 | 0.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.1 | GO:1904747 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.0 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.0 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.0 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.0 | 0.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.0 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.0 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.0 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.0 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.0 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.1 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.0 | 0.1 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.0 | GO:1903912 | regulation of translation in response to endoplasmic reticulum stress(GO:0036490) regulation of translation initiation in response to endoplasmic reticulum stress(GO:0036491) eiF2alpha phosphorylation in response to endoplasmic reticulum stress(GO:0036492) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.0 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.1 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.2 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.0 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.0 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.1 | 0.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.2 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.3 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0034843 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.0 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.3 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0015927 | trehalase activity(GO:0015927) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.0 | 0.0 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.0 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0043910 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 1.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |