Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
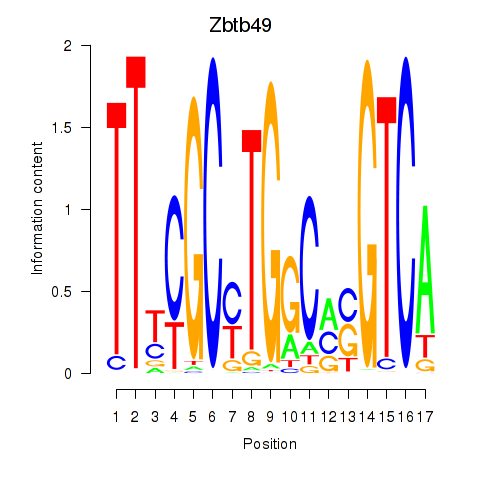
Results for Zbtb49
Z-value: 0.67
Transcription factors associated with Zbtb49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb49
|
ENSMUSG00000029127.9 | zinc finger and BTB domain containing 49 |
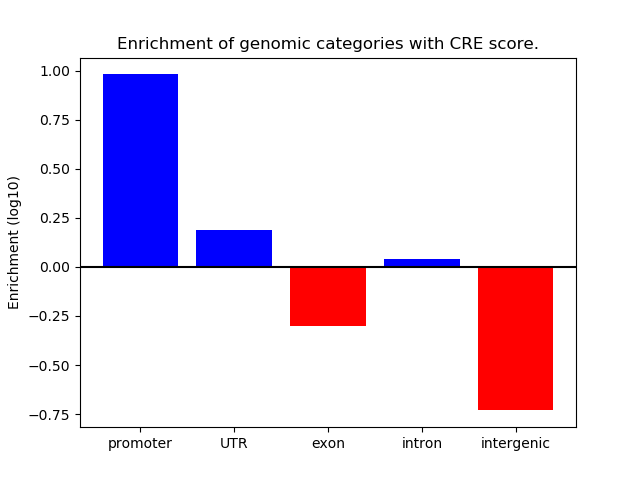
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_38219951_38220398 | Zbtb49 | 239 | 0.551365 | 0.09 | 4.8e-01 | Click! |
| chr5_38218735_38218938 | Zbtb49 | 1577 | 0.269187 | -0.04 | 7.7e-01 | Click! |
Activity of the Zbtb49 motif across conditions
Conditions sorted by the z-value of the Zbtb49 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
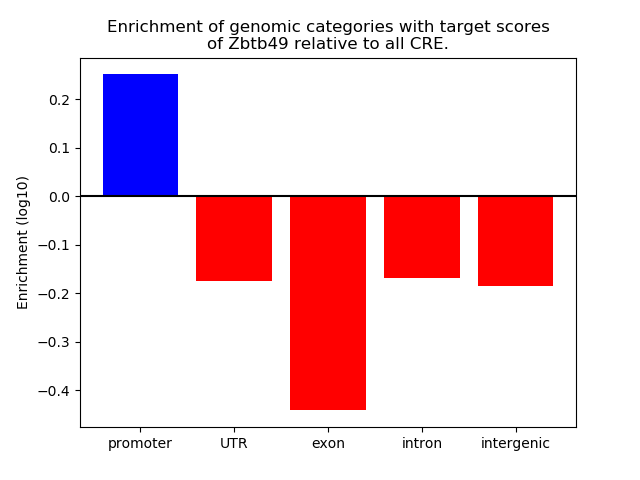
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_115908055_115909691 | 2.28 |
Cit |
citron |
1403 |
0.37 |
| chr12_61525659_61526870 | 2.11 |
Lrfn5 |
leucine rich repeat and fibronectin type III domain containing 5 |
2316 |
0.3 |
| chr9_96728847_96729194 | 2.06 |
Zbtb38 |
zinc finger and BTB domain containing 38 |
16 |
0.97 |
| chr3_4798346_4798833 | 1.82 |
1110015O18Rik |
RIKEN cDNA 1110015O18 gene |
119 |
0.97 |
| chr8_84769068_84769679 | 1.69 |
Nfix |
nuclear factor I/X |
4023 |
0.13 |
| chr7_140154045_140155011 | 1.56 |
Sprn |
shadow of prion protein |
349 |
0.75 |
| chr2_181155937_181157234 | 1.51 |
Eef1a2 |
eukaryotic translation elongation factor 1 alpha 2 |
429 |
0.73 |
| chr9_40268795_40270233 | 1.32 |
Scn3b |
sodium channel, voltage-gated, type III, beta |
81 |
0.96 |
| chr7_126822762_126823312 | 1.28 |
Fam57b |
family with sequence similarity 57, member B |
266 |
0.74 |
| chr5_5264770_5266186 | 1.18 |
Cdk14 |
cyclin-dependent kinase 14 |
169 |
0.96 |
| chr2_92399780_92399931 | 1.16 |
Mapk8ip1 |
mitogen-activated protein kinase 8 interacting protein 1 |
1408 |
0.25 |
| chr13_8207058_8207829 | 1.14 |
Adarb2 |
adenosine deaminase, RNA-specific, B2 |
4521 |
0.2 |
| chr4_24429901_24430719 | 1.13 |
Gm27243 |
predicted gene 27243 |
580 |
0.79 |
| chrX_103184741_103184892 | 1.08 |
Nap1l2 |
nucleosome assembly protein 1-like 2 |
1824 |
0.25 |
| chr16_43510268_43510725 | 1.06 |
Zbtb20 |
zinc finger and BTB domain containing 20 |
188 |
0.96 |
| chr1_32172319_32173236 | 1.04 |
Khdrbs2 |
KH domain containing, RNA binding, signal transduction associated 2 |
29 |
0.99 |
| chr1_164454554_164455430 | 0.99 |
Atp1b1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
1215 |
0.39 |
| chr7_139833633_139836105 | 0.99 |
Adgra1 |
adhesion G protein-coupled receptor A1 |
93 |
0.96 |
| chr10_127165174_127166341 | 0.92 |
B4galnt1 |
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
74 |
0.93 |
| chr9_113792658_113793898 | 0.91 |
Clasp2 |
CLIP associating protein 2 |
48 |
0.98 |
| chr19_39812576_39812727 | 0.87 |
Cyp2c40 |
cytochrome P450, family 2, subfamily c, polypeptide 40 |
93 |
0.98 |
| chr15_76519928_76521866 | 0.87 |
Scrt1 |
scratch family zinc finger 1 |
1005 |
0.28 |
| chr1_83407298_83408547 | 0.87 |
Sphkap |
SPHK1 interactor, AKAP domain containing |
217 |
0.94 |
| chr18_61910602_61911795 | 0.84 |
Ablim3 |
actin binding LIM protein family, member 3 |
625 |
0.73 |
| chr1_132199896_132201879 | 0.82 |
Lemd1 |
LEM domain containing 1 |
76 |
0.95 |
| chr8_95703051_95704038 | 0.80 |
Ndrg4 |
N-myc downstream regulated gene 4 |
474 |
0.68 |
| chr13_69736281_69736725 | 0.80 |
Ube2ql1 |
ubiquitin-conjugating enzyme E2Q family-like 1 |
3386 |
0.16 |
| chr5_120428678_120429275 | 0.80 |
Lhx5 |
LIM homeobox protein 5 |
2723 |
0.18 |
| chr4_122955098_122956090 | 0.80 |
Mfsd2a |
major facilitator superfamily domain containing 2A |
4396 |
0.16 |
| chr18_25747291_25747951 | 0.78 |
Celf4 |
CUGBP, Elav-like family member 4 |
5071 |
0.25 |
| chr12_36153893_36154044 | 0.76 |
Bzw2 |
basic leucine zipper and W2 domains 2 |
2659 |
0.17 |
| chr5_112576144_112577196 | 0.76 |
Sez6l |
seizure related 6 homolog like |
198 |
0.92 |
| chr8_4492894_4494304 | 0.75 |
Cers4 |
ceramide synthase 4 |
23 |
0.97 |
| chr10_104819342_104819493 | 0.75 |
Gm25522 |
predicted gene, 25522 |
45891 |
0.17 |
| chr5_70379643_70379794 | 0.68 |
Gm26072 |
predicted gene, 26072 |
171229 |
0.03 |
| chr9_21037316_21038046 | 0.68 |
Gm26592 |
predicted gene, 26592 |
101 |
0.9 |
| chr8_120492701_120493230 | 0.68 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
4518 |
0.17 |
| chr5_27261637_27262396 | 0.67 |
Dpp6 |
dipeptidylpeptidase 6 |
41 |
0.98 |
| chrX_136666183_136667703 | 0.66 |
Tceal3 |
transcription elongation factor A (SII)-like 3 |
332 |
0.84 |
| chr12_80769848_80770025 | 0.65 |
Ccdc177 |
coiled-coil domain containing 177 |
9249 |
0.12 |
| chr17_24643558_24645911 | 0.65 |
Slc9a3r2 |
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
223 |
0.82 |
| chr5_137553079_137554278 | 0.65 |
Actl6b |
actin-like 6B |
121 |
0.89 |
| chr5_24607000_24607197 | 0.64 |
Smarcd3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
5086 |
0.11 |
| chr15_76721855_76722516 | 0.63 |
Lrrc24 |
leucine rich repeat containing 24 |
12 |
0.92 |
| chr15_74565280_74565930 | 0.61 |
Adgrb1 |
adhesion G protein-coupled receptor B1 |
1704 |
0.27 |
| chr11_96305580_96306179 | 0.61 |
Hoxb5os |
homeobox B5 and homeobox B6, opposite strand |
798 |
0.33 |
| chr7_25607189_25607340 | 0.60 |
Gm4607 |
predicted gene 4607 |
5938 |
0.09 |
| chr2_106699616_106699779 | 0.60 |
Mpped2 |
metallophosphoesterase domain containing 2 |
3843 |
0.29 |
| chr2_115898028_115898179 | 0.59 |
Meis2 |
Meis homeobox 2 |
25141 |
0.25 |
| chr4_127015642_127016128 | 0.59 |
Sfpq |
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
5439 |
0.13 |
| chr11_73281234_73281630 | 0.58 |
Trpv3 |
transient receptor potential cation channel, subfamily V, member 3 |
14044 |
0.1 |
| chr10_18470218_18471289 | 0.57 |
Nhsl1 |
NHS-like 1 |
772 |
0.72 |
| chr3_60489739_60489921 | 0.57 |
Mbnl1 |
muscleblind like splicing factor 1 |
11105 |
0.24 |
| chr18_64340633_64341751 | 0.56 |
Onecut2 |
one cut domain, family member 2 |
1172 |
0.45 |
| chr1_119525279_119525430 | 0.56 |
Tmem185b |
transmembrane protein 185B |
806 |
0.38 |
| chr11_109653199_109653966 | 0.56 |
Prkar1a |
protein kinase, cAMP dependent regulatory, type I, alpha |
2661 |
0.23 |
| chr1_109982396_109983459 | 0.55 |
Cdh7 |
cadherin 7, type 2 |
54 |
0.99 |
| chr15_103011882_103012844 | 0.55 |
Hoxc5 |
homeobox C5 |
1452 |
0.19 |
| chr14_96517868_96518996 | 0.55 |
Klhl1 |
kelch-like 1 |
670 |
0.78 |
| chr1_87184698_87185439 | 0.55 |
Prss56 |
protease, serine 56 |
1755 |
0.2 |
| chr5_115475784_115476389 | 0.54 |
Sirt4 |
sirtuin 4 |
3852 |
0.1 |
| chr4_119539113_119539523 | 0.53 |
Frg2f1 |
FSHD region gene 2 family member 1 |
188 |
0.65 |
| chr6_137252157_137253458 | 0.52 |
Ptpro |
protein tyrosine phosphatase, receptor type, O |
342 |
0.92 |
| chr15_34495344_34495864 | 0.51 |
Pop1 |
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
281 |
0.54 |
| chr6_28924063_28924220 | 0.51 |
Snd1 |
staphylococcal nuclease and tudor domain containing 1 |
44098 |
0.13 |
| chr19_5691875_5693863 | 0.51 |
Map3k11 |
mitogen-activated protein kinase kinase kinase 11 |
26 |
0.92 |
| chr9_54286058_54287026 | 0.50 |
Gldn |
gliomedin |
56 |
0.97 |
| chr2_30358021_30359257 | 0.50 |
Sh3glb2 |
SH3-domain GRB2-like endophilin B2 |
601 |
0.55 |
| chr10_107890113_107890543 | 0.50 |
Otogl |
otogelin-like |
21806 |
0.24 |
| chr9_50856443_50856925 | 0.49 |
Ppp2r1b |
protein phosphatase 2, regulatory subunit A, beta |
240 |
0.91 |
| chr11_76396066_76396459 | 0.48 |
Nxn |
nucleoredoxin |
2806 |
0.25 |
| chr18_69384485_69384729 | 0.47 |
Tcf4 |
transcription factor 4 |
31290 |
0.22 |
| chr5_81021302_81022876 | 0.47 |
Adgrl3 |
adhesion G protein-coupled receptor L3 |
279 |
0.95 |
| chr11_105364219_105365174 | 0.46 |
Gm11638 |
predicted gene 11638 |
1444 |
0.39 |
| chr9_45432313_45432910 | 0.46 |
4833428L15Rik |
RIKEN cDNA 4833428L15 gene |
881 |
0.47 |
| chr4_148130089_148131325 | 0.46 |
Draxin |
dorsal inhibitory axon guidance protein |
9 |
0.72 |
| chr3_87959939_87961181 | 0.46 |
Gm3745 |
predicted gene 3745 |
7626 |
0.09 |
| chr11_23124548_23125169 | 0.45 |
1700061J23Rik |
RIKEN cDNA 1700061J23 gene |
27282 |
0.14 |
| chr4_134400063_134400411 | 0.45 |
Pafah2 |
platelet-activating factor acetylhydrolase 2 |
2675 |
0.18 |
| chr3_98941003_98941978 | 0.45 |
5730437C11Rik |
RIKEN cDNA 5730437C11 gene |
234 |
0.92 |
| chr17_44082773_44083036 | 0.45 |
Enpp5 |
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
4025 |
0.24 |
| chr15_57975359_57975957 | 0.45 |
Fam83a |
family with sequence similarity 83, member A |
9761 |
0.16 |
| chr7_16131445_16132800 | 0.44 |
Slc8a2 |
solute carrier family 8 (sodium/calcium exchanger), member 2 |
1774 |
0.23 |
| chr9_118227365_118228117 | 0.44 |
Gm17399 |
predicted gene, 17399 |
77510 |
0.09 |
| chr2_154570084_154570341 | 0.43 |
E2f1 |
E2F transcription factor 1 |
320 |
0.81 |
| chr17_13682256_13683548 | 0.43 |
Gm16046 |
predicted gene 16046 |
585 |
0.68 |
| chr13_99512095_99512261 | 0.42 |
Map1b |
microtubule-associated protein 1B |
4340 |
0.18 |
| chr15_94197097_94197248 | 0.42 |
Adamts20 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 20 |
207086 |
0.02 |
| chr4_82344703_82345124 | 0.42 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
94497 |
0.08 |
| chr7_141122888_141123988 | 0.41 |
Ptdss2 |
phosphatidylserine synthase 2 |
1056 |
0.29 |
| chr4_141052695_141054311 | 0.41 |
Crocc |
ciliary rootlet coiled-coil, rootletin |
158 |
0.92 |
| chr10_12940976_12941914 | 0.41 |
B230208H11Rik |
RIKEN cDNA B230208H11 gene |
18355 |
0.17 |
| chr2_154359544_154359723 | 0.41 |
Cdk5rap1 |
CDK5 regulatory subunit associated protein 1 |
5289 |
0.19 |
| chr18_47648563_47648791 | 0.41 |
Gm5236 |
predicted gene 5236 |
81090 |
0.08 |
| chr6_119328752_119331284 | 0.41 |
Lrtm2 |
leucine-rich repeats and transmembrane domains 2 |
739 |
0.43 |
| chr10_108365083_108365548 | 0.40 |
Gm23105 |
predicted gene, 23105 |
1650 |
0.39 |
| chr9_120305155_120305306 | 0.40 |
Myrip |
myosin VIIA and Rab interacting protein |
1294 |
0.38 |
| chr10_84060694_84061226 | 0.40 |
Gm37908 |
predicted gene, 37908 |
798 |
0.64 |
| chr13_102811110_102811952 | 0.40 |
Mast4 |
microtubule associated serine/threonine kinase family member 4 |
10610 |
0.23 |
| chr19_10041589_10042653 | 0.40 |
Fads3 |
fatty acid desaturase 3 |
389 |
0.78 |
| chr5_77460063_77460528 | 0.40 |
1700017L05Rik |
RIKEN cDNA 1700017L05 gene |
5193 |
0.17 |
| chr18_73863569_73864469 | 0.39 |
Mro |
maestro |
347 |
0.9 |
| chr5_125532320_125533519 | 0.39 |
Tmem132b |
transmembrane protein 132B |
532 |
0.76 |
| chr13_94025180_94025378 | 0.38 |
Cycs-ps3 |
cytochrome c, pseudogene 3 |
28564 |
0.14 |
| chr9_40453562_40453959 | 0.38 |
Gramd1b |
GRAM domain containing 1B |
1910 |
0.3 |
| chr4_101508533_101509308 | 0.38 |
Dnajc6 |
DnaJ heat shock protein family (Hsp40) member C6 |
890 |
0.64 |
| chr11_119913855_119915054 | 0.38 |
Chmp6 |
charged multivesicular body protein 6 |
439 |
0.76 |
| chr2_118862035_118862506 | 0.38 |
Ivd |
isovaleryl coenzyme A dehydrogenase |
242 |
0.89 |
| chr1_91398918_91399115 | 0.38 |
Ilkap |
integrin-linked kinase-associated serine/threonine phosphatase 2C |
201 |
0.89 |
| chr16_91646595_91646873 | 0.37 |
Gart |
phosphoribosylglycinamide formyltransferase |
178 |
0.84 |
| chr12_24657601_24657860 | 0.37 |
Klf11 |
Kruppel-like factor 11 |
5576 |
0.16 |
| chr7_42592149_42592824 | 0.37 |
Zfp977 |
zinc finger protein 977 |
61 |
0.96 |
| chr12_112722274_112723056 | 0.37 |
Cep170b |
centrosomal protein 170B |
471 |
0.67 |
| chr7_46099780_46100565 | 0.37 |
Kcnj11 |
potassium inwardly rectifying channel, subfamily J, member 11 |
65 |
0.95 |
| chr13_43284609_43284796 | 0.37 |
Gfod1 |
glucose-fructose oxidoreductase domain containing 1 |
18703 |
0.2 |
| chrX_8205722_8206558 | 0.36 |
Porcn |
porcupine O-acyltransferase |
351 |
0.79 |
| chr9_21130674_21131701 | 0.36 |
Tyk2 |
tyrosine kinase 2 |
1 |
0.95 |
| chr16_33847136_33847287 | 0.36 |
Itgb5 |
integrin beta 5 |
17534 |
0.16 |
| chr6_135526679_135526846 | 0.36 |
Gm25136 |
predicted gene, 25136 |
57430 |
0.13 |
| chr7_4993272_4994998 | 0.36 |
Zfp579 |
zinc finger protein 579 |
1205 |
0.21 |
| chr5_3151730_3152673 | 0.35 |
Gm8715 |
predicted gene 8715 |
28424 |
0.13 |
| chr5_143009425_143009646 | 0.35 |
Rnf216 |
ring finger protein 216 |
7239 |
0.16 |
| chr14_64126114_64126329 | 0.35 |
9630015K15Rik |
RIKEN cDNA 9630015K15 gene |
9907 |
0.13 |
| chr17_74316487_74316684 | 0.35 |
Dpy30 |
dpy-30, histone methyltransferase complex regulatory subunit |
122 |
0.94 |
| chr1_38835637_38836715 | 0.34 |
Lonrf2 |
LON peptidase N-terminal domain and ring finger 2 |
198 |
0.94 |
| chr17_6489757_6490685 | 0.34 |
Tmem181b-ps |
transmembrane protein 181B, pseudogene |
23199 |
0.14 |
| chr9_98428203_98428354 | 0.34 |
Rbp1 |
retinol binding protein 1, cellular |
5317 |
0.23 |
| chr15_63780688_63780839 | 0.34 |
Gm46499 |
predicted gene, 46499 |
20602 |
0.1 |
| chr7_142757390_142757797 | 0.33 |
Gm7290 |
predicted pseudogene 7290 |
259 |
0.9 |
| chr7_70929709_70930345 | 0.33 |
Gm34783 |
predicted gene, 34783 |
17207 |
0.23 |
| chr2_137163627_137163778 | 0.33 |
Gm28214 |
predicted gene 28214 |
35281 |
0.19 |
| chr14_34569383_34570378 | 0.33 |
Ldb3 |
LIM domain binding 3 |
7114 |
0.13 |
| chr13_94680868_94681019 | 0.33 |
Gm32305 |
predicted gene, 32305 |
19413 |
0.14 |
| chrX_9199674_9200884 | 0.33 |
Lancl3 |
LanC lantibiotic synthetase component C-like 3 (bacterial) |
377 |
0.83 |
| chr5_35448122_35448615 | 0.33 |
Gm43377 |
predicted gene 43377 |
52270 |
0.09 |
| chr11_72962601_72963125 | 0.33 |
Atp2a3 |
ATPase, Ca++ transporting, ubiquitous |
1573 |
0.28 |
| chr12_79191005_79191867 | 0.32 |
Rdh11 |
retinol dehydrogenase 11 |
357 |
0.78 |
| chrX_161719845_161720465 | 0.32 |
Rai2 |
retinoic acid induced 2 |
2528 |
0.41 |
| chr11_79340125_79340687 | 0.32 |
Nf1 |
neurofibromin 1 |
519 |
0.76 |
| chr14_33444094_33444245 | 0.32 |
Mapk8 |
mitogen-activated protein kinase 8 |
2973 |
0.2 |
| chr7_111054563_111054714 | 0.31 |
Ctr9 |
CTR9 homolog, Paf1/RNA polymerase II complex component |
848 |
0.54 |
| chr17_25809082_25810175 | 0.31 |
Fbxl16 |
F-box and leucine-rich repeat protein 16 |
543 |
0.42 |
| chr19_4126003_4127028 | 0.31 |
Tmem134 |
transmembrane protein 134 |
486 |
0.4 |
| chr17_6320795_6321910 | 0.31 |
AC183097.1 |
transmembrane protein 181 (TMEM181) pseudogene |
384 |
0.76 |
| chr11_97812280_97813382 | 0.31 |
Lasp1 |
LIM and SH3 protein 1 |
2417 |
0.14 |
| chr17_24208374_24209710 | 0.31 |
BC028777 |
cDNA sequence BC028777 |
181 |
0.66 |
| chr17_6256598_6257723 | 0.31 |
Tmem181a |
transmembrane protein 181A |
210 |
0.91 |
| chr3_142559923_142560770 | 0.31 |
Gbp3 |
guanylate binding protein 3 |
65 |
0.96 |
| chr12_75701139_75702422 | 0.30 |
Wdr89 |
WD repeat domain 89 |
32243 |
0.16 |
| chr5_38598055_38598428 | 0.30 |
4930421P07Rik |
RIKEN cDNA 4930421P07 gene |
23210 |
0.16 |
| chr1_186555744_186555937 | 0.29 |
A730004F24Rik |
RIKEN cDNA A730004F24 gene |
2632 |
0.36 |
| chr3_8684903_8685755 | 0.29 |
Gm23670 |
predicted gene, 23670 |
5852 |
0.16 |
| chr11_116030567_116031392 | 0.29 |
Unk |
unkempt family zinc finger |
656 |
0.53 |
| chr4_28812623_28812910 | 0.29 |
Epha7 |
Eph receptor A7 |
365 |
0.88 |
| chr7_73654506_73654727 | 0.28 |
Gm26176 |
predicted gene, 26176 |
8527 |
0.09 |
| chr2_28840464_28841222 | 0.28 |
Ddx31 |
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 31 |
384 |
0.54 |
| chr16_52450441_52451342 | 0.28 |
Alcam |
activated leukocyte cell adhesion molecule |
1574 |
0.55 |
| chr17_29883165_29884481 | 0.28 |
Mdga1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
3659 |
0.2 |
| chr3_58987083_58987709 | 0.28 |
Rpl13-ps6 |
ribosomal protein L13, pseudogene 6 |
226 |
0.9 |
| chr7_126702563_126704731 | 0.28 |
Coro1a |
coronin, actin binding protein 1A |
473 |
0.55 |
| chr2_127070718_127071288 | 0.28 |
Blvra |
biliverdin reductase A |
310 |
0.87 |
| chr3_94954464_94954835 | 0.28 |
Rfx5 |
regulatory factor X, 5 (influences HLA class II expression) |
114 |
0.92 |
| chr10_61080083_61080288 | 0.28 |
Pcbd1 |
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1 |
9146 |
0.13 |
| chr2_70126636_70126952 | 0.27 |
Myo3b |
myosin IIIB |
30496 |
0.2 |
| chr15_102203042_102203419 | 0.27 |
Zfp740 |
zinc finger protein 740 |
19 |
0.69 |
| chr1_87467212_87467595 | 0.27 |
Snorc |
secondary ossification center associated regulator of chondrocyte maturation |
2868 |
0.18 |
| chr2_58565057_58565662 | 0.26 |
Acvr1 |
activin A receptor, type 1 |
1467 |
0.38 |
| chr6_72598511_72599213 | 0.26 |
Retsat |
retinol saturase (all trans retinol 13,14 reductase) |
234 |
0.65 |
| chr3_51234712_51234887 | 0.26 |
Gm38357 |
predicted gene, 38357 |
2882 |
0.19 |
| chr3_127705323_127706456 | 0.26 |
1500005C15Rik |
RIKEN cDNA 1500005C15 gene |
11138 |
0.11 |
| chr8_25976746_25977095 | 0.26 |
Hgsnat |
heparan-alpha-glucosaminide N-acetyltransferase |
167 |
0.93 |
| chr12_58212274_58214477 | 0.26 |
Sstr1 |
somatostatin receptor 1 |
1571 |
0.52 |
| chr12_105456394_105456761 | 0.26 |
D430019H16Rik |
RIKEN cDNA D430019H16 gene |
2721 |
0.25 |
| chr11_79590775_79591929 | 0.26 |
Rab11fip4 |
RAB11 family interacting protein 4 (class II) |
140 |
0.93 |
| chr19_4282321_4283738 | 0.25 |
Ankrd13d |
ankyrin repeat domain 13 family, member D |
4 |
0.94 |
| chr5_138760327_138760734 | 0.25 |
Fam20c |
family with sequence similarity 20, member C |
5443 |
0.2 |
| chr11_72441409_72442226 | 0.25 |
Mybbp1a |
MYB binding protein (P160) 1a |
440 |
0.75 |
| chr2_25355573_25356319 | 0.25 |
Dpp7 |
dipeptidylpeptidase 7 |
112 |
0.9 |
| chr19_10769714_10769865 | 0.25 |
A430093F15Rik |
RIKEN cDNA A430093F15 gene |
12126 |
0.12 |
| chr3_106684672_106685318 | 0.25 |
Lrif1 |
ligand dependent nuclear receptor interacting factor 1 |
3 |
0.98 |
| chr7_123025258_123025409 | 0.25 |
Gm45846 |
predicted gene 45846 |
6041 |
0.14 |
| chr3_66292264_66292972 | 0.24 |
Veph1 |
ventricular zone expressed PH domain-containing 1 |
4130 |
0.27 |
| chrX_101532846_101533913 | 0.24 |
Taf1 |
TATA-box binding protein associated factor 1 |
620 |
0.69 |
| chr12_106080982_106081684 | 0.24 |
Gm46378 |
predicted gene, 46378 |
3037 |
0.25 |
| chr6_67037798_67037949 | 0.24 |
Gadd45a |
growth arrest and DNA-damage-inducible 45 alpha |
416 |
0.69 |
| chr17_24892273_24893815 | 0.24 |
Eme2 |
essential meiotic structure-specific endonuclease subunit 2 |
1141 |
0.23 |
| chr2_25400245_25401217 | 0.24 |
Npdc1 |
neural proliferation, differentiation and control 1 |
262 |
0.77 |
| chr17_79230829_79231720 | 0.24 |
Gm5230 |
predicted gene 5230 |
26775 |
0.21 |
| chr19_15193718_15194239 | 0.24 |
Gm5513 |
predicted pseudogene 5513 |
233212 |
0.02 |
| chr17_24736672_24737401 | 0.24 |
Msrb1 |
methionine sulfoxide reductase B1 |
354 |
0.65 |
| chr9_83834063_83834549 | 0.24 |
Ttk |
Ttk protein kinase |
383 |
0.87 |
| chr19_34192022_34193121 | 0.23 |
Stambpl1 |
STAM binding protein like 1 |
251 |
0.92 |
| chr9_107553913_107554411 | 0.23 |
Rassf1 |
Ras association (RalGDS/AF-6) domain family member 1 |
421 |
0.54 |
| chr2_118558327_118558555 | 0.23 |
Bmf |
BCL2 modifying factor |
8754 |
0.16 |
| chr10_95291310_95291461 | 0.23 |
Gm48880 |
predicted gene, 48880 |
23468 |
0.13 |
| chr15_77894951_77895251 | 0.23 |
Txn2 |
thioredoxin 2 |
20599 |
0.13 |
| chr17_86964856_86965294 | 0.23 |
Rhoq |
ras homolog family member Q |
1783 |
0.25 |
| chr4_130519685_130520343 | 0.23 |
Nkain1 |
Na+/K+ transporting ATPase interacting 1 |
37974 |
0.17 |
| chr6_59208973_59209748 | 0.23 |
Tigd2 |
tigger transposable element derived 2 |
490 |
0.83 |
| chr16_65819058_65819209 | 0.23 |
Vgll3 |
vestigial like family member 3 |
3504 |
0.29 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 1.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.2 | 0.9 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.4 | GO:1903286 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.3 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.1 | 0.4 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.1 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.2 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.2 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.1 | 0.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.6 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.1 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.4 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.3 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.2 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.4 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 0.9 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.2 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 1.6 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 0.6 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.5 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:1990035 | calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.3 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.0 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.1 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.5 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 1.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.4 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.3 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.3 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 1.2 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.0 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 0.0 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.5 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:0090151 | establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 2.0 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.1 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0097466 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.0 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.0 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.0 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.0 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:1903795 | regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.0 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.0 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 0.0 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 0.9 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 1.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 1.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.3 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.6 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.4 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.5 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.4 | GO:0052890 | oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.0 | 0.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.2 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.5 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.0 | REACTOME SIGNALLING TO ERKS | Genes involved in Signalling to ERKs |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |