Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
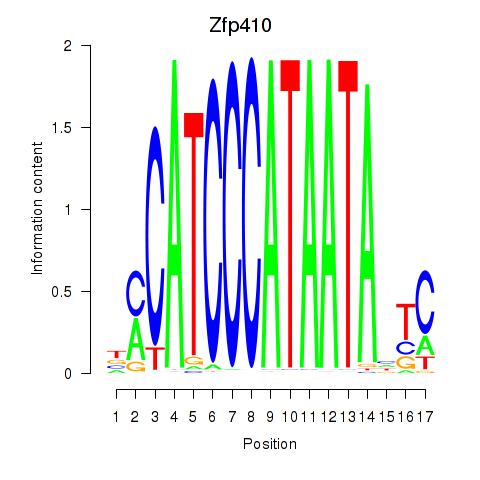
Results for Zfp410
Z-value: 0.66
Transcription factors associated with Zfp410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp410
|
ENSMUSG00000042472.10 | zinc finger protein 410 |
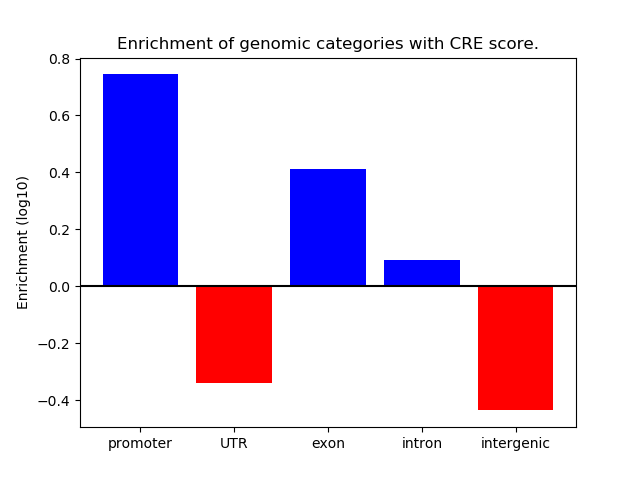
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_84316195_84316346 | Zfp410 | 582 | 0.477805 | -0.38 | 2.4e-03 | Click! |
| chr12_84318817_84318968 | Zfp410 | 1747 | 0.208145 | 0.29 | 2.3e-02 | Click! |
| chr12_84319037_84319188 | Zfp410 | 1967 | 0.191599 | 0.29 | 2.3e-02 | Click! |
| chr12_84319266_84319417 | Zfp410 | 2196 | 0.178225 | 0.28 | 3.1e-02 | Click! |
| chr12_84318564_84318715 | Zfp410 | 1494 | 0.233369 | 0.22 | 8.7e-02 | Click! |
Activity of the Zfp410 motif across conditions
Conditions sorted by the z-value of the Zfp410 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
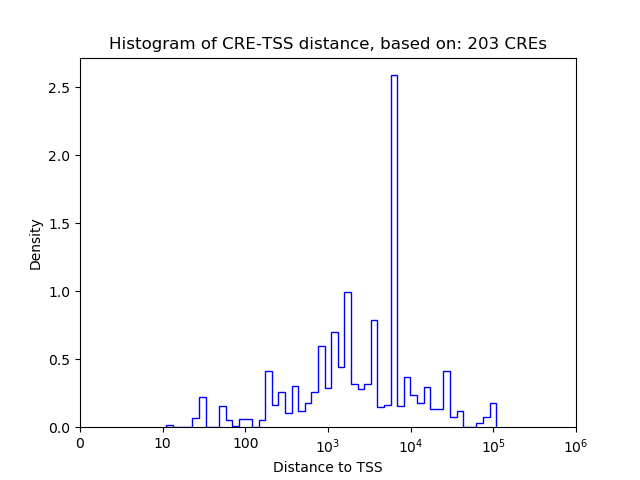
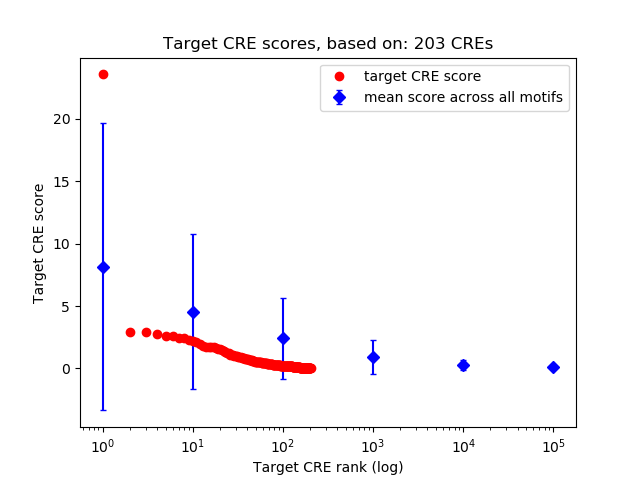
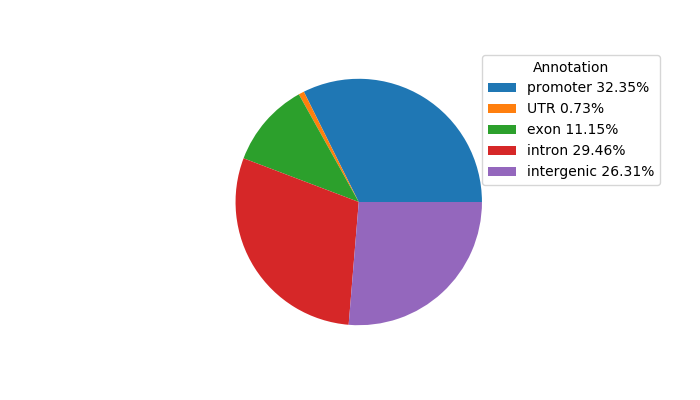
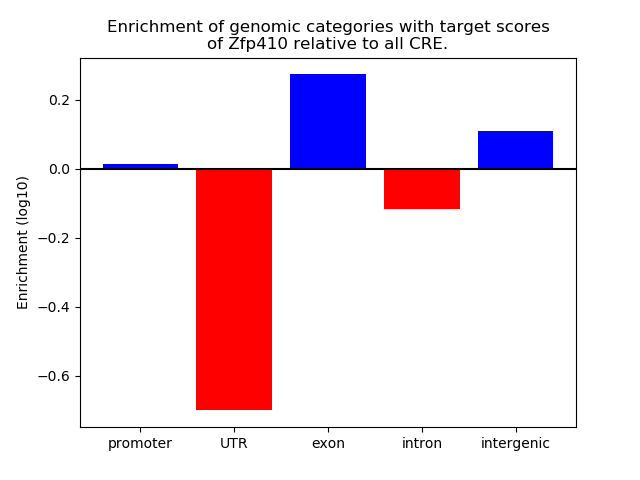
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_125088526_125090642 | 23.56 |
2010008C14Rik |
RIKEN cDNA 2010008C14 gene |
6147 |
0.07 |
| chr1_174173964_174174767 | 2.92 |
Spta1 |
spectrin alpha, erythrocytic 1 |
1589 |
0.22 |
| chr16_22893216_22893797 | 2.90 |
Gm30505 |
predicted gene, 30505 |
880 |
0.37 |
| chr4_32986761_32986927 | 2.72 |
Rragd |
Ras-related GTP binding D |
3390 |
0.16 |
| chr15_78261783_78262576 | 2.59 |
Ncf4 |
neutrophil cytosolic factor 4 |
1301 |
0.34 |
| chr2_31006029_31006516 | 2.57 |
Usp20 |
ubiquitin specific peptidase 20 |
3711 |
0.18 |
| chr10_60077941_60078202 | 2.47 |
Spock2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
28148 |
0.16 |
| chr10_56106927_56107500 | 2.41 |
Msl3l2 |
MSL3 like 2 |
296 |
0.92 |
| chr9_111146491_111146642 | 2.24 |
Lrrfip2 |
leucine rich repeat (in FLII) interacting protein 2 |
14737 |
0.16 |
| chr1_184729496_184731200 | 2.16 |
Hlx |
H2.0-like homeobox |
1250 |
0.37 |
| chr4_123284834_123285406 | 2.13 |
Pabpc4 |
poly(A) binding protein, cytoplasmic 4 |
2025 |
0.17 |
| chr2_32386034_32386242 | 1.98 |
Lcn2 |
lipocalin 2 |
31 |
0.94 |
| chr4_154924143_154924717 | 1.82 |
Tnfrsf14 |
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) |
3647 |
0.15 |
| chr1_131525650_131527377 | 1.73 |
Srgap2 |
SLIT-ROBO Rho GTPase activating protein 2 |
834 |
0.47 |
| chr6_41700699_41701150 | 1.72 |
Kel |
Kell blood group |
1756 |
0.24 |
| chr2_131454053_131454421 | 1.70 |
Gm14304 |
predicted gene 14304 |
403 |
0.82 |
| chr5_103763438_103763619 | 1.69 |
Aff1 |
AF4/FMR2 family, member 1 |
8955 |
0.21 |
| chr16_26368806_26369157 | 1.63 |
Cldn1 |
claudin 1 |
2860 |
0.38 |
| chr13_37581137_37581365 | 1.58 |
Gm47754 |
predicted gene, 47754 |
174 |
0.87 |
| chr14_52196647_52197712 | 1.54 |
Supt16 |
SPT16, facilitates chromatin remodeling subunit |
237 |
0.56 |
| chr2_30658719_30659142 | 1.50 |
Gm14486 |
predicted gene 14486 |
10978 |
0.15 |
| chr6_52178790_52180823 | 1.39 |
5730596B20Rik |
RIKEN cDNA 5730596B20 gene |
2308 |
0.09 |
| chr1_174171570_174172092 | 1.30 |
Spta1 |
spectrin alpha, erythrocytic 1 |
945 |
0.37 |
| chr7_4747526_4747677 | 1.24 |
Kmt5c |
lysine methyltransferase 5C |
1793 |
0.15 |
| chr6_67013848_67014074 | 1.20 |
Gm15644 |
predicted gene 15644 |
1476 |
0.27 |
| chr5_31887658_31887897 | 1.11 |
Gm43811 |
predicted gene 43811 |
38920 |
0.16 |
| chr15_102102931_102104191 | 1.10 |
Tns2 |
tensin 2 |
573 |
0.62 |
| chr14_21178664_21178815 | 1.04 |
Adk |
adenosine kinase |
102587 |
0.07 |
| chr5_28457868_28460972 | 1.03 |
9530036O11Rik |
RIKEN cDNA 9530036O11Rik |
1146 |
0.51 |
| chr5_100499385_100500654 | 1.02 |
Gm23222 |
predicted gene, 23222 |
199 |
0.78 |
| chr15_83169748_83171160 | 1.01 |
Cyb5r3 |
cytochrome b5 reductase 3 |
52 |
0.95 |
| chr4_108849566_108849717 | 0.91 |
Kti12 |
KTI12 homolog, chromatin associated |
1856 |
0.25 |
| chr15_100668768_100669476 | 0.91 |
Bin2 |
bridging integrator 2 |
383 |
0.73 |
| chr5_43515484_43516406 | 0.88 |
C1qtnf7 |
C1q and tumor necrosis factor related protein 7 |
183 |
0.94 |
| chr18_35555130_35555680 | 0.86 |
Snhg4 |
small nucleolar RNA host gene 4 |
734 |
0.29 |
| chr14_26441045_26441600 | 0.86 |
Slmap |
sarcolemma associated protein |
1333 |
0.39 |
| chr11_70639659_70639949 | 0.84 |
Gp1ba |
glycoprotein 1b, alpha polypeptide |
660 |
0.39 |
| chr11_102368401_102368706 | 0.79 |
Slc4a1 |
solute carrier family 4 (anion exchanger), member 1 |
2350 |
0.16 |
| chr18_74740823_74741114 | 0.77 |
Myo5b |
myosin VB |
7203 |
0.18 |
| chr2_130576014_130578077 | 0.75 |
Oxt |
oxytocin |
872 |
0.39 |
| chr13_5714127_5714791 | 0.74 |
Gm35330 |
predicted gene, 35330 |
9983 |
0.27 |
| chr19_10023046_10023689 | 0.67 |
Rab3il1 |
RAB3A interacting protein (rabin3)-like 1 |
5102 |
0.13 |
| chr7_74722253_74722404 | 0.66 |
Gm7726 |
predicted gene 7726 |
22472 |
0.21 |
| chr2_6128217_6128792 | 0.64 |
Proser2 |
proline and serine rich 2 |
1635 |
0.31 |
| chr18_56920684_56921182 | 0.63 |
Marchf3 |
membrane associated ring-CH-type finger 3 |
4582 |
0.22 |
| chr11_58961317_58962255 | 0.63 |
H3f4 |
H3.4 histone |
26 |
0.92 |
| chr3_96244511_96244662 | 0.60 |
H2ac18 |
H2A clustered histone 18 |
911 |
0.15 |
| chr8_108886846_108887288 | 0.57 |
Gm38318 |
predicted gene, 38318 |
19505 |
0.2 |
| chr12_71877272_71877457 | 0.54 |
Daam1 |
dishevelled associated activator of morphogenesis 1 |
12366 |
0.21 |
| chr19_23687455_23688235 | 0.54 |
Ptar1 |
protein prenyltransferase alpha subunit repeat containing 1 |
101 |
0.95 |
| chr8_36639144_36639295 | 0.53 |
Dlc1 |
deleted in liver cancer 1 |
25276 |
0.24 |
| chr5_16553245_16554447 | 0.50 |
Hgf |
hepatocyte growth factor |
60 |
0.98 |
| chr1_56099318_56099469 | 0.50 |
1700003I22Rik |
RIKEN cDNA 1700003I22 gene |
80370 |
0.11 |
| chr9_13827814_13828725 | 0.48 |
Fam76b |
family with sequence similarity 76, member B |
534 |
0.64 |
| chr3_31957151_31957302 | 0.47 |
Gm37834 |
predicted gene, 37834 |
2462 |
0.39 |
| chrX_150549256_150549407 | 0.47 |
Alas2 |
aminolevulinic acid synthase 2, erythroid |
1872 |
0.24 |
| chr3_146599870_146600091 | 0.47 |
Uox |
urate oxidase |
2814 |
0.18 |
| chr5_136810567_136811453 | 0.46 |
Col26a1 |
collagen, type XXVI, alpha 1 |
27299 |
0.15 |
| chr3_84005656_84006186 | 0.45 |
Tmem131l |
transmembrane 131 like |
34207 |
0.18 |
| chr5_137424316_137424467 | 0.45 |
Rpl36-ps9 |
ribosomal protein L36, pseudogene 9 |
8596 |
0.11 |
| chr17_29115479_29115785 | 0.42 |
Rab44 |
RAB44, member RAS oncogene family |
1451 |
0.24 |
| chr4_152041050_152041419 | 0.42 |
Nol9 |
nucleolar protein 9 |
1871 |
0.19 |
| chr17_24471773_24471948 | 0.41 |
Pgp |
phosphoglycolate phosphatase |
1425 |
0.15 |
| chr14_55194266_55194459 | 0.41 |
Gm46455 |
predicted gene, 46455 |
24653 |
0.1 |
| chr14_61257948_61258209 | 0.41 |
Sgcg |
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
346 |
0.85 |
| chr10_42268858_42269374 | 0.40 |
Foxo3 |
forkhead box O3 |
7580 |
0.26 |
| chr7_64040303_64040644 | 0.39 |
Gm45054 |
predicted gene 45054 |
353 |
0.85 |
| chr19_8929328_8930627 | 0.37 |
Eml3 |
echinoderm microtubule associated protein like 3 |
84 |
0.84 |
| chr4_109253812_109254165 | 0.35 |
Calr4 |
calreticulin 4 |
9406 |
0.19 |
| chr15_40674755_40675016 | 0.34 |
Zfpm2 |
zinc finger protein, multitype 2 |
9895 |
0.29 |
| chr4_87801519_87801738 | 0.33 |
Mllt3 |
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
4666 |
0.34 |
| chr2_153495481_153495632 | 0.33 |
4930404H24Rik |
RIKEN cDNA 4930404H24 gene |
2766 |
0.24 |
| chr3_59129690_59130148 | 0.33 |
P2ry14 |
purinergic receptor P2Y, G-protein coupled, 14 |
703 |
0.66 |
| chr7_104507498_104507806 | 0.33 |
Trim30d |
tripartite motif-containing 30D |
173 |
0.89 |
| chr1_91538647_91539044 | 0.32 |
Asb1 |
ankyrin repeat and SOCS box-containing 1 |
1699 |
0.3 |
| chrX_164421189_164421355 | 0.32 |
Piga |
phosphatidylinositol glycan anchor biosynthesis, class A |
1312 |
0.41 |
| chr9_99708109_99709014 | 0.31 |
Gm16004 |
predicted gene 16004 |
162 |
0.93 |
| chrX_52151061_52151857 | 0.31 |
Gpc4 |
glypican 4 |
13793 |
0.28 |
| chr1_36088938_36089276 | 0.30 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
12583 |
0.13 |
| chr18_38962190_38962341 | 0.30 |
Gm5820 |
predicted gene 5820 |
6556 |
0.19 |
| chr16_90916648_90916799 | 0.29 |
Gm7831 |
predicted gene 7831 |
1543 |
0.24 |
| chr1_156222209_156222829 | 0.29 |
Gm38113 |
predicted gene, 38113 |
4774 |
0.18 |
| chr18_77870000_77870716 | 0.29 |
Epg5 |
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
68107 |
0.09 |
| chr9_73044202_73045612 | 0.29 |
Rab27a |
RAB27A, member RAS oncogene family |
50 |
0.94 |
| chr16_13545571_13545722 | 0.28 |
Gm25276 |
predicted gene, 25276 |
14095 |
0.15 |
| chr5_145115755_145116764 | 0.28 |
Arpc1b |
actin related protein 2/3 complex, subunit 1B |
1943 |
0.18 |
| chr10_81351824_81352381 | 0.27 |
Hmg20b |
high mobility group 20B |
1622 |
0.13 |
| chr7_19159561_19160813 | 0.25 |
Gipr |
gastric inhibitory polypeptide receptor |
4907 |
0.08 |
| chr5_121233582_121234161 | 0.24 |
Hectd4 |
HECT domain E3 ubiquitin protein ligase 4 |
13652 |
0.13 |
| chr2_116768611_116768926 | 0.24 |
Gm13990 |
predicted gene 13990 |
108400 |
0.06 |
| chr14_54958124_54958655 | 0.24 |
Myh6 |
myosin, heavy polypeptide 6, cardiac muscle, alpha |
482 |
0.52 |
| chr17_35231958_35233195 | 0.24 |
Atp6v1g2 |
ATPase, H+ transporting, lysosomal V1 subunit G2 |
1084 |
0.18 |
| chr5_82246250_82246401 | 0.24 |
Rpl7-ps7 |
ribosomal protein L7, pseudogene 7 |
90526 |
0.09 |
| chr6_137630324_137630475 | 0.23 |
Eps8 |
epidermal growth factor receptor pathway substrate 8 |
17884 |
0.22 |
| chr14_59439369_59439520 | 0.23 |
Setdb2 |
SET domain, bifurcated 2 |
1429 |
0.3 |
| chr4_142166102_142166510 | 0.23 |
Kazn |
kazrin, periplakin interacting protein |
7252 |
0.19 |
| chr13_38635539_38635690 | 0.23 |
Bloc1s5 |
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
505 |
0.5 |
| chr5_114173960_114175108 | 0.22 |
Acacb |
acetyl-Coenzyme A carboxylase beta |
1372 |
0.35 |
| chr2_180273759_180274154 | 0.22 |
Cables2 |
CDK5 and Abl enzyme substrate 2 |
460 |
0.72 |
| chr17_65604240_65604425 | 0.22 |
Vapa |
vesicle-associated membrane protein, associated protein A |
9223 |
0.16 |
| chr2_26298030_26300214 | 0.21 |
Ccdc187 |
coiled-coil domain containing 187 |
4565 |
0.13 |
| chr6_29784657_29784847 | 0.21 |
Ahcyl2 |
S-adenosylhomocysteine hydrolase-like 2 |
14799 |
0.16 |
| chr16_5205031_5205182 | 0.21 |
Nagpa |
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
1094 |
0.33 |
| chr15_83166752_83167017 | 0.21 |
Cyb5r3 |
cytochrome b5 reductase 3 |
3293 |
0.13 |
| chr1_160906608_160907521 | 0.21 |
Rc3h1 |
RING CCCH (C3H) domains 1 |
646 |
0.5 |
| chr7_115860217_115860368 | 0.21 |
Sox6 |
SRY (sex determining region Y)-box 6 |
440 |
0.9 |
| chr17_47335038_47335189 | 0.20 |
Mrps10 |
mitochondrial ribosomal protein S10 |
33774 |
0.11 |
| chr16_21787099_21788407 | 0.20 |
Ehhadh |
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase |
54 |
0.96 |
| chr4_135405096_135405380 | 0.19 |
Gm12990 |
predicted gene 12990 |
1582 |
0.22 |
| chr6_117610478_117610687 | 0.19 |
Gm45083 |
predicted gene 45083 |
1988 |
0.36 |
| chr9_22155677_22155979 | 0.19 |
Pigyl |
phosphatidylinositol glycan anchor biosynthesis, class Y-like |
1018 |
0.29 |
| chr11_72458192_72459273 | 0.18 |
Spns2 |
spinster homolog 2 |
2111 |
0.22 |
| chr11_120169034_120170072 | 0.18 |
Slc38a10 |
solute carrier family 38, member 10 |
18207 |
0.09 |
| chr9_29288118_29288329 | 0.18 |
Ntm |
neurotrimin |
102558 |
0.08 |
| chr11_100964042_100964193 | 0.18 |
Cavin1 |
caveolae associated 1 |
6434 |
0.14 |
| chr3_19635362_19635513 | 0.18 |
1700064H15Rik |
RIKEN cDNA 1700064H15 gene |
6760 |
0.16 |
| chr5_123142167_123142508 | 0.18 |
Setd1b |
SET domain containing 1B |
144 |
0.83 |
| chr5_64715305_64715496 | 0.18 |
Gm20033 |
predicted gene, 20033 |
951 |
0.51 |
| chr8_75095618_75095913 | 0.18 |
Hmox1 |
heme oxygenase 1 |
1079 |
0.41 |
| chr2_9918739_9918949 | 0.17 |
Taf3 |
TATA-box binding protein associated factor 3 |
10 |
0.96 |
| chr15_36500071_36500348 | 0.17 |
Gm49246 |
predicted gene, 49246 |
3222 |
0.18 |
| chr5_147303089_147303425 | 0.17 |
Cdx2 |
caudal type homeobox 2 |
4013 |
0.12 |
| chr2_76655635_76655786 | 0.16 |
Pjvk |
pejvakin |
5437 |
0.16 |
| chr1_43162619_43162984 | 0.16 |
Fhl2 |
four and a half LIM domains 2 |
1160 |
0.46 |
| chr7_111691798_111691949 | 0.16 |
Galnt18 |
polypeptide N-acetylgalactosaminyltransferase 18 |
88104 |
0.09 |
| chr12_16731274_16731620 | 0.16 |
Greb1 |
gene regulated by estrogen in breast cancer protein |
8681 |
0.18 |
| chr11_118034790_118034941 | 0.15 |
Pgs1 |
phosphatidylglycerophosphate synthase 1 |
22522 |
0.14 |
| chr9_42097699_42097850 | 0.15 |
Sorl1 |
sortilin-related receptor, LDLR class A repeats-containing |
26523 |
0.17 |
| chr8_110933315_110933853 | 0.14 |
St3gal2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
1663 |
0.24 |
| chr8_35410929_35411173 | 0.14 |
Gm45301 |
predicted gene 45301 |
1545 |
0.35 |
| chr9_56572818_56573408 | 0.13 |
Gm47178 |
predicted gene, 47178 |
29357 |
0.13 |
| chr1_180847897_180848799 | 0.13 |
Sde2 |
SDE2 telomere maintenance homolog (S. pombe) |
2779 |
0.14 |
| chr4_138219311_138219462 | 0.13 |
Hp1bp3 |
heterochromatin protein 1, binding protein 3 |
2092 |
0.2 |
| chr5_140735262_140736522 | 0.13 |
Amz1 |
archaelysin family metallopeptidase 1 |
325 |
0.88 |
| chr3_58162274_58162814 | 0.13 |
1700007F19Rik |
RIKEN cDNA 1700007F19 gene |
1263 |
0.35 |
| chr9_84090334_84090732 | 0.13 |
Bckdhb |
branched chain ketoacid dehydrogenase E1, beta polypeptide |
16974 |
0.22 |
| chr15_76207208_76209116 | 0.13 |
Plec |
plectin |
91 |
0.92 |
| chr17_64469558_64469919 | 0.12 |
Mir6420 |
microRNA 6420 |
28932 |
0.22 |
| chr16_87681170_87681353 | 0.12 |
Bach1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
17684 |
0.2 |
| chr10_44476893_44477182 | 0.12 |
Prdm1 |
PR domain containing 1, with ZNF domain |
18289 |
0.18 |
| chr18_3279746_3279934 | 0.11 |
Crem |
cAMP responsive element modulator |
1238 |
0.52 |
| chr14_48133184_48133904 | 0.11 |
Gm49310 |
predicted gene, 49310 |
680 |
0.61 |
| chr7_128612386_128612537 | 0.11 |
Inpp5f |
inositol polyphosphate-5-phosphatase F |
1101 |
0.39 |
| chr17_29321890_29323144 | 0.10 |
Gm46603 |
predicted gene, 46603 |
32 |
0.95 |
| chr2_133556756_133557496 | 0.09 |
Bmp2 |
bone morphogenetic protein 2 |
4967 |
0.21 |
| chr10_12996822_12997284 | 0.09 |
Sf3b5 |
splicing factor 3b, subunit 5 |
8290 |
0.19 |
| chr6_36819200_36819780 | 0.09 |
Ptn |
pleiotrophin |
9270 |
0.28 |
| chr16_38714001_38714161 | 0.08 |
Arhgap31 |
Rho GTPase activating protein 31 |
807 |
0.56 |
| chr4_47437269_47438175 | 0.08 |
Gm12430 |
predicted gene 12430 |
15910 |
0.18 |
| chr11_95580318_95580469 | 0.08 |
Ngfr |
nerve growth factor receptor (TNFR superfamily, member 16) |
7342 |
0.17 |
| chr9_64734273_64734844 | 0.08 |
Rab11a |
RAB11A, member RAS oncogene family |
1953 |
0.32 |
| chr2_166081092_166081487 | 0.08 |
Sulf2 |
sulfatase 2 |
3170 |
0.24 |
| chr4_129331767_129331965 | 0.08 |
Rbbp4 |
retinoblastoma binding protein 4, chromatin remodeling factor |
2778 |
0.16 |
| chr5_77406870_77407021 | 0.07 |
Igfbp7 |
insulin-like growth factor binding protein 7 |
1095 |
0.43 |
| chr12_112588763_112589606 | 0.07 |
Inf2 |
inverted formin, FH2 and WH2 domain containing |
82 |
0.96 |
| chr11_87737570_87738432 | 0.07 |
Supt4a |
SPT4A, DSIF elongation factor subunit |
448 |
0.66 |
| chr6_126532364_126532878 | 0.07 |
Kcna5 |
potassium voltage-gated channel, shaker-related subfamily, member 5 |
2791 |
0.26 |
| chr4_41724852_41725139 | 0.07 |
Dctn3 |
dynactin 3 |
1825 |
0.18 |
| chr11_98202836_98204568 | 0.07 |
Cdk12 |
cyclin-dependent kinase 12 |
367 |
0.78 |
| chr9_106885777_106887024 | 0.07 |
Rbm15b |
RNA binding motif protein 15B |
781 |
0.48 |
| chr5_25705265_25705879 | 0.07 |
Xrcc2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
192 |
0.92 |
| chr3_60529662_60530216 | 0.07 |
Mbnl1 |
muscleblind like splicing factor 1 |
308 |
0.91 |
| chr3_66985349_66986398 | 0.07 |
Rsrc1 |
arginine/serine-rich coiled-coil 1 |
94 |
0.97 |
| chr11_108145810_108146207 | 0.06 |
Gm11655 |
predicted gene 11655 |
35842 |
0.19 |
| chr1_66944417_66944813 | 0.06 |
Myl1 |
myosin, light polypeptide 1 |
528 |
0.7 |
| chr10_80085003_80085473 | 0.06 |
Sbno2 |
strawberry notch 2 |
9799 |
0.09 |
| chrX_134717661_134719186 | 0.06 |
Armcx1 |
armadillo repeat containing, X-linked 1 |
403 |
0.72 |
| chr7_49778971_49779409 | 0.06 |
Prmt3 |
protein arginine N-methyltransferase 3 |
646 |
0.74 |
| chr5_131697484_131697635 | 0.06 |
Gm42442 |
predicted gene 42442 |
794 |
0.53 |
| chr8_106054482_106054922 | 0.06 |
Nfatc3 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
4138 |
0.12 |
| chr19_28677931_28680368 | 0.06 |
Glis3 |
GLIS family zinc finger 3 |
459 |
0.68 |
| chr13_13953757_13954320 | 0.06 |
B3galnt2 |
UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 2 |
431 |
0.62 |
| chr2_68466086_68466372 | 0.05 |
Stk39 |
serine/threonine kinase 39 |
5711 |
0.21 |
| chr17_5831890_5832283 | 0.05 |
Gm26622 |
predicted gene, 26622 |
5882 |
0.15 |
| chr9_77939183_77940182 | 0.05 |
Elovl5 |
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
1609 |
0.32 |
| chr11_110881943_110882098 | 0.05 |
Kcnj16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
86013 |
0.1 |
| chr5_124262016_124262210 | 0.05 |
Mphosph9 |
M-phase phosphoprotein 9 |
899 |
0.42 |
| chr4_116558201_116558717 | 0.05 |
Gpbp1l1 |
GC-rich promoter binding protein 1-like 1 |
380 |
0.75 |
| chr18_77766808_77767753 | 0.05 |
Haus1 |
HAUS augmin-like complex, subunit 1 |
500 |
0.72 |
| chr13_27325128_27325279 | 0.04 |
Gm11358 |
predicted gene 11358 |
6225 |
0.11 |
| chr2_157423895_157425112 | 0.04 |
Src |
Rous sarcoma oncogene |
111 |
0.97 |
| chr11_106329400_106329556 | 0.04 |
Cd79b |
CD79B antigen |
14716 |
0.1 |
| chr13_104226999_104227150 | 0.03 |
Cenpk |
centromere protein K |
1537 |
0.29 |
| chr7_25755347_25756122 | 0.03 |
Hnrnpul1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
977 |
0.37 |
| chr7_99590707_99590917 | 0.03 |
Arrb1 |
arrestin, beta 1 |
3781 |
0.14 |
| chr16_13545827_13545978 | 0.03 |
Gm25276 |
predicted gene, 25276 |
14351 |
0.15 |
| chr1_180726093_180726901 | 0.03 |
Acbd3 |
acyl-Coenzyme A binding domain containing 3 |
454 |
0.72 |
| chr13_83603651_83603866 | 0.03 |
Mef2c |
myocyte enhancer factor 2C |
21845 |
0.22 |
| chrX_73483606_73484999 | 0.03 |
Bgn |
biglycan |
663 |
0.59 |
| chr10_116871781_116872422 | 0.02 |
Myrfl |
myelin regulatory factor-like |
24818 |
0.15 |
| chr4_131829238_131829510 | 0.02 |
Ptpru |
protein tyrosine phosphatase, receptor type, U |
7858 |
0.15 |
| chr11_65421515_65421666 | 0.02 |
Gm12295 |
predicted gene 12295 |
55792 |
0.15 |
| chr7_90447549_90447852 | 0.02 |
Crebzf |
CREB/ATF bZIP transcription factor |
2997 |
0.17 |
| chrX_100782647_100782805 | 0.02 |
Dlg3 |
discs large MAGUK scaffold protein 3 |
5441 |
0.19 |
| chr4_134453606_134454237 | 0.01 |
1700021N21Rik |
RIKEN cDNA 1700021N21 gene |
3750 |
0.13 |
| chr10_61898411_61898967 | 0.01 |
Col13a1 |
collagen, type XIII, alpha 1 |
20297 |
0.19 |
| chr6_57531398_57531878 | 0.01 |
Ppm1k |
protein phosphatase 1K (PP2C domain containing) |
3788 |
0.19 |
| chr4_106827556_106828820 | 0.01 |
Gm12746 |
predicted gene 12746 |
19732 |
0.15 |
| chr8_95804942_95805303 | 0.01 |
4930513N10Rik |
RIKEN cDNA 4930513N10 gene |
1622 |
0.19 |
| chr16_13546060_13546211 | 0.01 |
Gm25276 |
predicted gene, 25276 |
14584 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.3 | 1.9 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.1 | 2.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.8 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.9 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.5 | GO:1902947 | regulation of tau-protein kinase activity(GO:1902947) |
| 0.1 | 1.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.3 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.1 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.4 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.1 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.3 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 0.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.2 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 | 0.8 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.1 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.4 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 | 0.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.3 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.1 | 1.3 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.7 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 1.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.5 | GO:0018119 | peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0002584 | negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.0 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.0 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.2 | GO:0045072 | regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0035898 | parathyroid hormone secretion(GO:0035898) |
| 0.0 | 0.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.0 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 2.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 1.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.9 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 1.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 1.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.7 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 1.2 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.6 | GO:1990003 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.0 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.0 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 1.0 | GO:0052735 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.0 | GO:0019198 | transmembrane receptor protein phosphatase activity(GO:0019198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |