Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
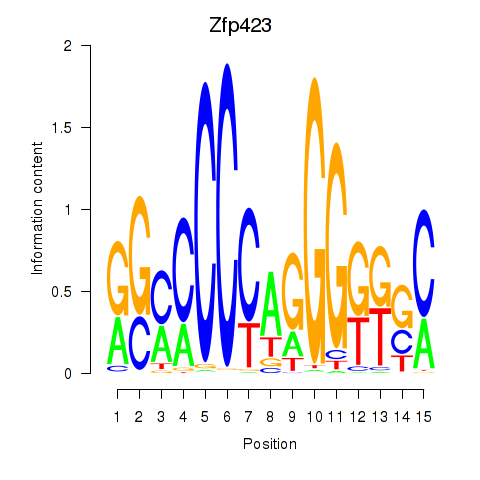
Results for Zfp423
Z-value: 0.72
Transcription factors associated with Zfp423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp423
|
ENSMUSG00000045333.9 | zinc finger protein 423 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_87803562_87804241 | Zfp423 | 78 | 0.984386 | -0.43 | 6.3e-04 | Click! |
| chr8_87955583_87955973 | Zfp423 | 1916 | 0.435773 | 0.33 | 9.8e-03 | Click! |
| chr8_87954422_87954717 | Zfp423 | 707 | 0.770329 | 0.33 | 1.1e-02 | Click! |
| chr8_87707568_87707784 | Zfp423 | 96147 | 0.074288 | 0.32 | 1.3e-02 | Click! |
| chr8_87955974_87956211 | Zfp423 | 2230 | 0.398047 | 0.30 | 1.9e-02 | Click! |
Activity of the Zfp423 motif across conditions
Conditions sorted by the z-value of the Zfp423 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_83724722_83725570 | 1.69 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
2960 |
0.17 |
| chr6_55454267_55454889 | 1.65 |
Adcyap1r1 |
adenylate cyclase activating polypeptide 1 receptor 1 |
2401 |
0.32 |
| chr1_17145379_17145927 | 1.41 |
Gdap1 |
ganglioside-induced differentiation-associated-protein 1 |
185 |
0.94 |
| chr6_55678280_55679200 | 1.38 |
Neurod6 |
neurogenic differentiation 6 |
2523 |
0.32 |
| chr2_180889143_180889758 | 1.28 |
Gm14342 |
predicted gene 14342 |
210 |
0.87 |
| chr8_84715808_84716438 | 1.20 |
Nfix |
nuclear factor I/X |
2055 |
0.18 |
| chr15_78116357_78116678 | 1.18 |
A730060N03Rik |
RIKEN cDNA A730060N03 gene |
3189 |
0.19 |
| chr15_80091999_80092465 | 1.14 |
Rpl3 |
ribosomal protein L3 |
364 |
0.65 |
| chr13_20473087_20474265 | 1.14 |
Gm32036 |
predicted gene, 32036 |
186 |
0.89 |
| chr6_110645148_110646464 | 1.12 |
Gm20387 |
predicted gene 20387 |
110 |
0.67 |
| chr1_42697532_42698715 | 1.10 |
Pou3f3 |
POU domain, class 3, transcription factor 3 |
2355 |
0.2 |
| chr8_94997776_94998724 | 1.09 |
Adgrg1 |
adhesion G protein-coupled receptor G1 |
2589 |
0.18 |
| chr3_114904046_114905354 | 1.07 |
Olfm3 |
olfactomedin 3 |
65 |
0.98 |
| chr4_46990051_46990797 | 0.92 |
Gabbr2 |
gamma-aminobutyric acid (GABA) B receptor, 2 |
1449 |
0.38 |
| chr7_30914787_30914938 | 0.88 |
Mag |
myelin-associated glycoprotein |
3 |
0.93 |
| chr7_29167642_29167990 | 0.86 |
Spred3 |
sprouty-related EVH1 domain containing 3 |
666 |
0.43 |
| chr15_64917271_64918226 | 0.85 |
Adcy8 |
adenylate cyclase 8 |
4523 |
0.29 |
| chr18_82405647_82407688 | 0.83 |
Galr1 |
galanin receptor 1 |
110 |
0.96 |
| chr2_118922726_118922877 | 0.82 |
Chst14 |
carbohydrate sulfotransferase 14 |
3695 |
0.18 |
| chr1_132192093_132192244 | 0.82 |
Gm29695 |
predicted gene, 29695 |
711 |
0.37 |
| chr5_22746211_22746970 | 0.77 |
A930003O13Rik |
RIKEN cDNA A930003O13 gene |
297 |
0.65 |
| chr5_120437495_120438815 | 0.76 |
Gm27199 |
predicted gene 27199 |
6388 |
0.13 |
| chr1_89583524_89583694 | 0.73 |
Agap1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
3373 |
0.23 |
| chr1_78193153_78194695 | 0.72 |
Pax3 |
paired box 3 |
2914 |
0.3 |
| chr11_33202043_33204837 | 0.71 |
Tlx3 |
T cell leukemia, homeobox 3 |
149 |
0.86 |
| chr5_48598987_48600332 | 0.70 |
Kcnip4 |
Kv channel interacting protein 4 |
23 |
0.97 |
| chr11_97567490_97567827 | 0.70 |
Srcin1 |
SRC kinase signaling inhibitor 1 |
6271 |
0.13 |
| chr13_48158648_48160003 | 0.70 |
Gm36346 |
predicted gene, 36346 |
77926 |
0.08 |
| chr11_5129447_5129810 | 0.68 |
Emid1 |
EMI domain containing 1 |
148 |
0.94 |
| chr11_97573801_97574201 | 0.68 |
Srcin1 |
SRC kinase signaling inhibitor 1 |
46 |
0.96 |
| chrX_111894411_111895700 | 0.68 |
Gm45194 |
predicted gene 45194 |
3479 |
0.27 |
| chr9_108824114_108825614 | 0.68 |
Gm35025 |
predicted gene, 35025 |
3 |
0.87 |
| chr1_38847863_38848426 | 0.67 |
Lonrf2 |
LON peptidase N-terminal domain and ring finger 2 |
11433 |
0.16 |
| chr5_30714946_30715457 | 0.66 |
Dpysl5 |
dihydropyrimidinase-like 5 |
3300 |
0.18 |
| chr5_115110373_115110642 | 0.66 |
Rpl37rt |
ribosomal protein L37, retrotransposed |
239 |
0.85 |
| chr1_191885619_191885770 | 0.65 |
1700034H15Rik |
RIKEN cDNA 1700034H15 gene |
13435 |
0.14 |
| chr14_52313293_52314443 | 0.65 |
Sall2 |
spalt like transcription factor 2 |
2455 |
0.13 |
| chr9_15874335_15874636 | 0.64 |
Mtnr1b |
melatonin receptor 1B |
25 |
0.98 |
| chr15_78769503_78769918 | 0.64 |
Mfng |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
2184 |
0.21 |
| chr8_34061456_34061607 | 0.63 |
Gm9951 |
predicted gene 9951 |
6909 |
0.14 |
| chr19_8949995_8950322 | 0.62 |
Tut1 |
terminal uridylyl transferase 1, U6 snRNA-specific |
3689 |
0.08 |
| chr1_92849002_92850443 | 0.61 |
Mir149 |
microRNA 149 |
656 |
0.43 |
| chr11_120238467_120239478 | 0.60 |
Bahcc1 |
BAH domain and coiled-coil containing 1 |
2273 |
0.16 |
| chr11_115091354_115092281 | 0.58 |
Rab37 |
RAB37, member RAS oncogene family |
386 |
0.77 |
| chr7_81488721_81488872 | 0.57 |
Ap3b2 |
adaptor-related protein complex 3, beta 2 subunit |
1014 |
0.38 |
| chr2_116063125_116063508 | 0.57 |
Meis2 |
Meis homeobox 2 |
786 |
0.59 |
| chr10_104456207_104456415 | 0.57 |
Gm22945 |
predicted gene, 22945 |
14896 |
0.27 |
| chr11_119725151_119725314 | 0.56 |
Gm23663 |
predicted gene, 23663 |
21169 |
0.18 |
| chr2_174074417_174075662 | 0.56 |
Stx16 |
syntaxin 16 |
1269 |
0.43 |
| chr18_64344305_64344524 | 0.55 |
Onecut2 |
one cut domain, family member 2 |
4394 |
0.19 |
| chr12_108395917_108396068 | 0.55 |
Eml1 |
echinoderm microtubule associated protein like 1 |
14550 |
0.17 |
| chr8_58912073_58913264 | 0.54 |
Galntl6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
28 |
0.88 |
| chr12_102555272_102555761 | 0.54 |
Chga |
chromogranin A |
530 |
0.74 |
| chr15_74563319_74564610 | 0.54 |
Adgrb1 |
adhesion G protein-coupled receptor B1 |
63 |
0.61 |
| chr8_126298317_126299213 | 0.53 |
Slc35f3 |
solute carrier family 35, member F3 |
207 |
0.96 |
| chr1_173351157_173351371 | 0.53 |
Aim2 |
absent in melanoma 2 |
385 |
0.82 |
| chr1_75263860_75264397 | 0.53 |
Ptprn |
protein tyrosine phosphatase, receptor type, N |
78 |
0.92 |
| chr8_48579238_48579993 | 0.53 |
Tenm3 |
teneurin transmembrane protein 3 |
23576 |
0.23 |
| chr12_86681553_86682494 | 0.53 |
Vash1 |
vasohibin 1 |
3323 |
0.18 |
| chr13_12105751_12107037 | 0.52 |
Ryr2 |
ryanodine receptor 2, cardiac |
65 |
0.97 |
| chr11_96368922_96369073 | 0.52 |
Hoxb1 |
homeobox B1 |
2739 |
0.13 |
| chr13_116302578_116304501 | 0.52 |
Isl1 |
ISL1 transcription factor, LIM/homeodomain |
188 |
0.96 |
| chr13_65420114_65420941 | 0.52 |
Cbx3-ps5 |
chromobox 3, pseudogene 5 |
3926 |
0.09 |
| chr4_153316698_153317280 | 0.52 |
Gm13174 |
predicted gene 13174 |
98813 |
0.08 |
| chr13_65562682_65563537 | 0.51 |
Cbx3-ps2 |
chromobox 3, pseudogene 2 |
3941 |
0.1 |
| chr15_44787203_44788294 | 0.51 |
A930017M01Rik |
RIKEN cDNA A930017M01 gene |
21 |
0.83 |
| chr19_5659277_5659564 | 0.51 |
Sipa1 |
signal-induced proliferation associated gene 1 |
2215 |
0.12 |
| chr9_45430098_45431532 | 0.50 |
4833428L15Rik |
RIKEN cDNA 4833428L15 gene |
417 |
0.52 |
| chr17_55986138_55987186 | 0.50 |
Fsd1 |
fibronectin type 3 and SPRY domain-containing protein |
147 |
0.89 |
| chr11_96343236_96346574 | 0.50 |
Hoxb3 |
homeobox B3 |
1136 |
0.24 |
| chr17_34547078_34547229 | 0.49 |
Btnl7-ps |
butyrophilin-like 7, pseudogene |
498 |
0.54 |
| chr15_74516560_74519631 | 0.49 |
Adgrb1 |
adhesion G protein-coupled receptor B1 |
1264 |
0.47 |
| chr10_81399790_81400870 | 0.49 |
Nfic |
nuclear factor I/C |
224 |
0.78 |
| chr14_39473185_39473855 | 0.49 |
Nrg3 |
neuregulin 3 |
432 |
0.91 |
| chr17_56682332_56682605 | 0.49 |
Ranbp3 |
RAN binding protein 3 |
9084 |
0.11 |
| chr8_50915347_50916555 | 0.48 |
C130073E24Rik |
RIKEN cDNA C130073E24 gene |
1046 |
0.7 |
| chr13_45849543_45849787 | 0.48 |
Atxn1 |
ataxin 1 |
22623 |
0.23 |
| chr11_82508511_82509165 | 0.48 |
Tmem132e |
transmembrane protein 132E |
70581 |
0.09 |
| chr5_137734408_137735271 | 0.47 |
Nyap1 |
neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 |
4879 |
0.1 |
| chrX_170674573_170675954 | 0.47 |
Asmt |
acetylserotonin O-methyltransferase |
2619 |
0.41 |
| chr1_36088252_36088532 | 0.46 |
Hs6st1 |
heparan sulfate 6-O-sulfotransferase 1 |
13298 |
0.13 |
| chr19_44670373_44671507 | 0.46 |
Gm26644 |
predicted gene, 26644 |
24335 |
0.14 |
| chr16_18428270_18428753 | 0.46 |
Txnrd2 |
thioredoxin reductase 2 |
186 |
0.88 |
| chr9_121426905_121427083 | 0.45 |
Trak1 |
trafficking protein, kinesin binding 1 |
11019 |
0.19 |
| chr19_6375769_6375920 | 0.45 |
Sf1 |
splicing factor 1 |
1715 |
0.17 |
| chr14_54228777_54229582 | 0.45 |
Traj1 |
T cell receptor alpha joining 1 |
10365 |
0.08 |
| chr14_122280336_122281519 | 0.45 |
Clybl |
citrate lyase beta like |
22886 |
0.18 |
| chr1_59376471_59376724 | 0.45 |
Gm29016 |
predicted gene 29016 |
2333 |
0.31 |
| chr7_37389133_37389284 | 0.45 |
6720469O03Rik |
RIKEN cDNA 6720469O03 gene |
22578 |
0.21 |
| chr10_80663215_80663381 | 0.44 |
Btbd2 |
BTB (POZ) domain containing 2 |
6227 |
0.09 |
| chr19_8949777_8949945 | 0.43 |
Mta2 |
metastasis-associated gene family, member 2 |
3461 |
0.08 |
| chr8_92350062_92350844 | 0.43 |
Crnde |
colorectal neoplasia differentially expressed (non-protein coding) |
3583 |
0.22 |
| chr9_108189365_108190636 | 0.43 |
Gm37247 |
predicted gene, 37247 |
273 |
0.55 |
| chr13_65351676_65352548 | 0.42 |
Gm7762 |
predicted gene 7762 |
3515 |
0.09 |
| chr4_72195085_72196381 | 0.42 |
Tle1 |
transducin-like enhancer of split 1 |
3359 |
0.26 |
| chr5_117492934_117494252 | 0.42 |
Gm42550 |
predicted gene 42550 |
27106 |
0.18 |
| chr5_3929547_3929947 | 0.42 |
Akap9 |
A kinase (PRKA) anchor protein (yotiao) 9 |
1473 |
0.38 |
| chr11_96365680_96367114 | 0.41 |
Hoxb1 |
homeobox B1 |
139 |
0.91 |
| chr1_172318526_172318677 | 0.41 |
Igsf8 |
immunoglobulin superfamily, member 8 |
159 |
0.91 |
| chr12_53251739_53251890 | 0.41 |
Npas3 |
neuronal PAS domain protein 3 |
1211 |
0.61 |
| chr3_88537757_88539123 | 0.41 |
Mir1905 |
microRNA 1905 |
2058 |
0.13 |
| chr12_33928070_33929277 | 0.41 |
Gm40383 |
predicted gene, 40383 |
217 |
0.53 |
| chr1_182764585_182764988 | 0.41 |
Susd4 |
sushi domain containing 4 |
109 |
0.97 |
| chr14_72531887_72532977 | 0.41 |
Mlnr-ps |
motilin receptor, pseudogene |
217 |
0.94 |
| chr8_123885000_123885457 | 0.41 |
Acta1 |
actin, alpha 1, skeletal muscle |
9523 |
0.09 |
| chr5_107238027_107238178 | 0.41 |
Gm8145 |
predicted gene 8145 |
3521 |
0.19 |
| chr6_83141558_83143227 | 0.41 |
Rtkn |
rhotekin |
0 |
0.92 |
| chr8_34815025_34816588 | 0.41 |
Dusp4 |
dual specificity phosphatase 4 |
8509 |
0.22 |
| chr11_116919762_116920015 | 0.40 |
Mgat5b |
mannoside acetylglucosaminyltransferase 5, isoenzyme B |
1025 |
0.48 |
| chr2_179536834_179536985 | 0.40 |
Gm14300 |
predicted gene 14300 |
76658 |
0.1 |
| chr1_92737523_92737805 | 0.40 |
Gm29483 |
predicted gene 29483 |
20462 |
0.12 |
| chr13_65890336_65891205 | 0.40 |
Cbx3-ps1 |
chromobox 3, pseudogene 1 |
3989 |
0.13 |
| chr8_119832782_119833285 | 0.39 |
Cotl1 |
coactosin-like 1 (Dictyostelium) |
7464 |
0.17 |
| chr9_54502671_54503218 | 0.39 |
Dmxl2 |
Dmx-like 2 |
1318 |
0.46 |
| chr4_136954751_136954947 | 0.39 |
Epha8 |
Eph receptor A8 |
1967 |
0.29 |
| chr6_37900496_37900647 | 0.39 |
Trim24 |
tripartite motif-containing 24 |
84 |
0.98 |
| chr2_151968587_151968817 | 0.39 |
Mir1953 |
microRNA 1953 |
1085 |
0.4 |
| chr13_35615438_35615768 | 0.38 |
Gm48707 |
predicted gene, 48707 |
601 |
0.76 |
| chr8_12390154_12391412 | 0.38 |
Gm45560 |
predicted gene 45560 |
615 |
0.63 |
| chr2_104097569_104098085 | 0.38 |
Cd59a |
CD59a antigen |
1987 |
0.22 |
| chr8_4216103_4217573 | 0.38 |
Prr36 |
proline rich 36 |
74 |
0.93 |
| chr5_120433178_120434996 | 0.38 |
Gm27199 |
predicted gene 27199 |
2320 |
0.19 |
| chr12_3806578_3808221 | 0.37 |
Dnmt3a |
DNA methyltransferase 3A |
239 |
0.92 |
| chr5_142778290_142778581 | 0.37 |
Tnrc18 |
trinucleotide repeat containing 18 |
10088 |
0.19 |
| chr7_45870455_45871042 | 0.36 |
Grin2d |
glutamate receptor, ionotropic, NMDA2D (epsilon 4) |
319 |
0.72 |
| chr1_164454554_164455430 | 0.36 |
Atp1b1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
1215 |
0.39 |
| chr9_32340821_32341078 | 0.36 |
Kcnj5 |
potassium inwardly-rectifying channel, subfamily J, member 5 |
3288 |
0.22 |
| chr5_110486743_110486894 | 0.36 |
Gm32996 |
predicted gene, 32996 |
4617 |
0.16 |
| chr19_37233614_37234686 | 0.36 |
Gm25268 |
predicted gene, 25268 |
666 |
0.62 |
| chr19_6501834_6503156 | 0.36 |
Nrxn2 |
neurexin II |
4660 |
0.14 |
| chr3_88206822_88208169 | 0.36 |
Gm3764 |
predicted gene 3764 |
183 |
0.86 |
| chr19_10199751_10200167 | 0.36 |
Gm10143 |
predicted gene 10143 |
770 |
0.44 |
| chr9_78377654_78378912 | 0.36 |
Ooep |
oocyte expressed protein |
442 |
0.65 |
| chr15_39197166_39197982 | 0.35 |
Rims2 |
regulating synaptic membrane exocytosis 2 |
687 |
0.66 |
| chr13_66867979_66868968 | 0.35 |
Cbx3-ps4 |
chromobox 3, pseudogene 4 |
3949 |
0.09 |
| chr7_16435189_16435340 | 0.35 |
Tmem160 |
transmembrane protein 160 |
17515 |
0.09 |
| chr2_92983089_92983290 | 0.35 |
Prdm11 |
PR domain containing 11 |
30721 |
0.15 |
| chr10_80380857_80382709 | 0.35 |
Mex3d |
mex3 RNA binding family member D |
204 |
0.7 |
| chr8_70674781_70675218 | 0.35 |
Lsm4 |
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
1567 |
0.19 |
| chr13_116301327_116301857 | 0.35 |
Isl1 |
ISL1 transcription factor, LIM/homeodomain |
1759 |
0.42 |
| chr16_91222072_91224159 | 0.35 |
Gm49614 |
predicted gene, 49614 |
189 |
0.9 |
| chrX_100769299_100769740 | 0.35 |
Dlg3 |
discs large MAGUK scaffold protein 3 |
1524 |
0.34 |
| chr17_15373239_15373914 | 0.35 |
Dll1 |
delta like canonical Notch ligand 1 |
496 |
0.76 |
| chr1_66467636_66468234 | 0.35 |
Unc80 |
unc-80, NALCN activator |
432 |
0.85 |
| chr17_57069929_57071109 | 0.35 |
Gm49888 |
predicted gene, 49888 |
1159 |
0.21 |
| chr9_69767833_69768898 | 0.34 |
B230323A14Rik |
RIKEN cDNA B230323A14 gene |
7219 |
0.19 |
| chr12_117152536_117153210 | 0.34 |
Gm10421 |
predicted gene 10421 |
1222 |
0.62 |
| chr9_91379793_91379944 | 0.34 |
Zic4 |
zinc finger protein of the cerebellum 4 |
1226 |
0.29 |
| chr15_79667169_79667426 | 0.34 |
Tomm22 |
translocase of outer mitochondrial membrane 22 |
3564 |
0.11 |
| chr2_122366307_122366748 | 0.34 |
Shf |
Src homology 2 domain containing F |
301 |
0.86 |
| chr7_3652751_3653338 | 0.34 |
Cnot3 |
CCR4-NOT transcription complex, subunit 3 |
1473 |
0.16 |
| chr15_78468579_78468730 | 0.34 |
Tmprss6 |
transmembrane serine protease 6 |
20 |
0.96 |
| chr7_25271053_25271214 | 0.34 |
Cic |
capicua transcriptional repressor |
2746 |
0.12 |
| chr6_31587504_31587730 | 0.34 |
Gm6117 |
predicted gene 6117 |
12650 |
0.17 |
| chr5_122045490_122045656 | 0.34 |
Cux2 |
cut-like homeobox 2 |
2252 |
0.22 |
| chr7_25089847_25089998 | 0.34 |
Mir6537 |
microRNA 6537 |
7253 |
0.11 |
| chr11_88332695_88332846 | 0.33 |
Gm11509 |
predicted gene 11509 |
2890 |
0.21 |
| chr7_45008093_45008451 | 0.33 |
Mir7054 |
microRNA 7054 |
4215 |
0.07 |
| chr11_50228536_50229225 | 0.33 |
Mgat4b |
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
1860 |
0.19 |
| chr4_138447570_138447898 | 0.33 |
Camk2n1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
6580 |
0.14 |
| chr7_126950022_126951260 | 0.33 |
Sez6l2 |
seizure related 6 homolog like 2 |
57 |
0.89 |
| chr4_120649231_120650449 | 0.33 |
Gm45579 |
predicted gene 45579 |
1703 |
0.3 |
| chr3_34662808_34665047 | 0.33 |
Gm42693 |
predicted gene 42693 |
362 |
0.74 |
| chr2_22621739_22621917 | 0.33 |
Gad2 |
glutamic acid decarboxylase 2 |
377 |
0.81 |
| chr14_32599245_32600743 | 0.32 |
Prrxl1 |
paired related homeobox protein-like 1 |
36 |
0.97 |
| chr9_74853776_74854743 | 0.32 |
Gm28182 |
predicted gene 28182 |
1126 |
0.41 |
| chr19_6272278_6272429 | 0.32 |
Gm14963 |
predicted gene 14963 |
4252 |
0.08 |
| chr2_154632073_154632660 | 0.32 |
Gm14198 |
predicted gene 14198 |
270 |
0.85 |
| chr12_111420504_111420734 | 0.32 |
Exoc3l4 |
exocyst complex component 3-like 4 |
30 |
0.96 |
| chr8_12430022_12431770 | 0.32 |
Gm25239 |
predicted gene, 25239 |
34493 |
0.1 |
| chr11_18870055_18872175 | 0.32 |
8430419K02Rik |
RIKEN cDNA 8430419K02 gene |
1040 |
0.49 |
| chr18_36517168_36517319 | 0.32 |
Hbegf |
heparin-binding EGF-like growth factor |
1438 |
0.31 |
| chr12_98573488_98573793 | 0.31 |
Kcnk10 |
potassium channel, subfamily K, member 10 |
1072 |
0.43 |
| chr13_34340668_34341142 | 0.31 |
Slc22a23 |
solute carrier family 22, member 23 |
3298 |
0.23 |
| chr7_45867785_45869036 | 0.31 |
Grin2d |
glutamate receptor, ionotropic, NMDA2D (epsilon 4) |
1281 |
0.21 |
| chr9_54661035_54661331 | 0.31 |
Acsbg1 |
acyl-CoA synthetase bubblegum family member 1 |
512 |
0.74 |
| chr2_152091566_152091717 | 0.31 |
Scrt2 |
scratch family zinc finger 2 |
10112 |
0.13 |
| chr15_76519928_76521866 | 0.31 |
Scrt1 |
scratch family zinc finger 1 |
1005 |
0.28 |
| chr15_89532557_89533956 | 0.30 |
Shank3 |
SH3 and multiple ankyrin repeat domains 3 |
366 |
0.78 |
| chr4_126310931_126311082 | 0.30 |
Adprhl2 |
ADP-ribosylhydrolase like 2 |
10355 |
0.11 |
| chr4_148593276_148593471 | 0.30 |
Srm |
spermidine synthase |
521 |
0.62 |
| chr2_147189265_147191223 | 0.30 |
6430503K07Rik |
RIKEN cDNA 6430503K07 gene |
2820 |
0.2 |
| chr17_56142590_56142902 | 0.30 |
Sema6b |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
2403 |
0.14 |
| chr7_98817595_98817746 | 0.30 |
Wnt11 |
wingless-type MMTV integration site family, member 11 |
17442 |
0.15 |
| chrX_51017522_51018698 | 0.30 |
Rap2c |
RAP2C, member of RAS oncogene family |
92 |
0.98 |
| chr15_81990475_81990958 | 0.30 |
Xrcc6 |
X-ray repair complementing defective repair in Chinese hamster cells 6 |
2881 |
0.12 |
| chr5_136990060_136990211 | 0.30 |
Plod3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
171 |
0.85 |
| chr14_25127903_25128556 | 0.30 |
Gm32224 |
predicted gene, 32224 |
441 |
0.8 |
| chr7_83894733_83894884 | 0.30 |
Mesd |
mesoderm development LRP chaperone |
245 |
0.86 |
| chr17_56241459_56242409 | 0.30 |
A230051N06Rik |
RIKEN cDNA A230051N06 gene |
218 |
0.81 |
| chr8_122749580_122750238 | 0.30 |
C230057M02Rik |
RIKEN cDNA C230057M02 gene |
2416 |
0.16 |
| chr7_144896076_144897460 | 0.30 |
Fgf15 |
fibroblast growth factor 15 |
204 |
0.8 |
| chr9_110947230_110947482 | 0.29 |
Tdgf1 |
teratocarcinoma-derived growth factor 1 |
1198 |
0.27 |
| chr19_8594121_8594701 | 0.29 |
Slc22a8 |
solute carrier family 22 (organic anion transporter), member 8 |
3122 |
0.15 |
| chr7_139905381_139905970 | 0.29 |
Kndc1 |
kinase non-catalytic C-lobe domain (KIND) containing 1 |
10836 |
0.11 |
| chr13_99446279_99447668 | 0.29 |
Map1b |
microtubule-associated protein 1B |
647 |
0.72 |
| chr13_112439365_112440345 | 0.29 |
Gm48879 |
predicted gene, 48879 |
3259 |
0.19 |
| chr12_86084451_86084602 | 0.29 |
Ift43 |
intraflagellar transport 43 |
1918 |
0.26 |
| chr9_102353147_102354512 | 0.29 |
Ephb1 |
Eph receptor B1 |
864 |
0.59 |
| chr15_89085762_89086162 | 0.29 |
Trabd |
TraB domain containing |
1253 |
0.25 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 0.8 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.2 | 0.5 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.2 | 0.5 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.2 | 0.2 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.6 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.4 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.1 | 0.5 | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.1 | 0.4 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.6 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.4 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.1 | 0.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.3 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.1 | 0.3 | GO:0072240 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.2 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 0.8 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.1 | 0.4 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.2 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.2 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.4 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.3 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.7 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.2 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.1 | 0.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.2 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.1 | 0.1 | GO:0002339 | B cell selection(GO:0002339) |
| 0.1 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.4 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 1.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.6 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.2 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.2 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 | 0.2 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 0.7 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.1 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.1 | 0.2 | GO:0030167 | proteoglycan catabolic process(GO:0030167) |
| 0.0 | 0.1 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.3 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.3 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.0 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.3 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.0 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.9 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.1 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.4 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.0 | GO:0050883 | medulla oblongata development(GO:0021550) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0021557 | oculomotor nerve development(GO:0021557) trochlear nerve development(GO:0021558) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.4 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.0 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.0 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.2 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.0 | 0.3 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.3 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.7 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 | 0.3 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.1 | GO:0060926 | cardiac pacemaker cell development(GO:0060926) sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.6 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.0 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.0 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0021892 | cerebral cortex GABAergic interneuron differentiation(GO:0021892) |
| 0.0 | 0.1 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.0 | GO:0003057 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) regulation of the force of heart contraction by chemical signal(GO:0003057) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.0 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.0 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.0 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.0 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.2 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.0 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.1 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.1 | GO:0034982 | protein processing involved in protein targeting to mitochondrion(GO:0006627) mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.0 | 0.2 | GO:0031280 | negative regulation of cyclase activity(GO:0031280) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.0 | GO:0051462 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) |
| 0.0 | 0.2 | GO:1903859 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.0 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.0 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.0 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.0 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.1 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.0 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.8 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.0 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.0 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.0 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:1902308 | regulation of peptidyl-serine dephosphorylation(GO:1902308) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.2 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.3 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.2 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 0.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.0 | GO:1902965 | protein localization to early endosome(GO:1902946) regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.0 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.0 | GO:0061184 | positive regulation of dermatome development(GO:0061184) |
| 0.0 | 0.1 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.1 | GO:1900094 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0009068 | aspartate family amino acid catabolic process(GO:0009068) |
| 0.0 | 0.0 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.0 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.0 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.0 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.0 | GO:0009139 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.0 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.0 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.0 | 0.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.0 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.0 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.0 | GO:2000847 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.0 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.0 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.1 | GO:0006563 | L-serine metabolic process(GO:0006563) |
| 0.0 | 0.0 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.0 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.0 | GO:1904528 | regulation of microtubule binding(GO:1904526) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.4 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.0 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.0 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.0 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 0.4 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.3 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.0 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.9 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.0 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.1 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 0.8 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 0.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 0.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.2 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.4 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.3 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.2 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 1.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 1.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0001030 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.7 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.0 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 1.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.1 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |