Project
ENCODE: H3K4me3 ChIP-Seq of different mouse tissues during embryonic development
Navigation
Downloads
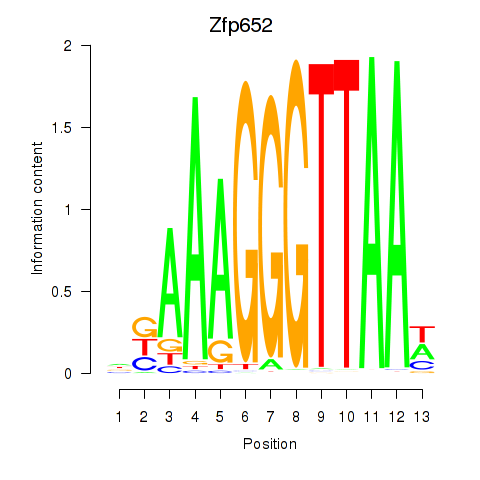
Results for Zfp652
Z-value: 1.27
Transcription factors associated with Zfp652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp652
|
ENSMUSG00000075595.3 | zinc finger protein 652 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_95716253_95716475 | Zfp652 | 3353 | 0.157796 | 0.53 | 1.5e-05 | Click! |
| chr11_95729019_95729298 | Zfp652 | 16147 | 0.116523 | -0.41 | 1.2e-03 | Click! |
| chr11_95713175_95714166 | Zfp652 | 659 | 0.467319 | 0.33 | 1.0e-02 | Click! |
| chr11_95708065_95708216 | Zfp652 | 4533 | 0.140730 | -0.07 | 5.7e-01 | Click! |
| chr11_95729627_95729778 | Zfp652 | 16691 | 0.115825 | 0.02 | 8.6e-01 | Click! |
Activity of the Zfp652 motif across conditions
Conditions sorted by the z-value of the Zfp652 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
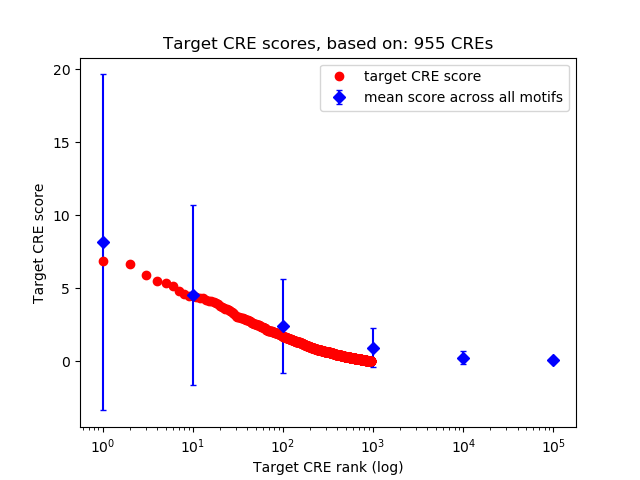
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_102352095_102352639 | 6.84 |
Ephb1 |
Eph receptor B1 |
2326 |
0.28 |
| chr14_64591036_64591686 | 6.68 |
Mir3078 |
microRNA 3078 |
176 |
0.84 |
| chr1_160350716_160351791 | 5.92 |
Rabgap1l |
RAB GTPase activating protein 1-like |
318 |
0.88 |
| chr14_52009953_52011160 | 5.52 |
Zfp219 |
zinc finger protein 219 |
19 |
0.94 |
| chr5_121008454_121009154 | 5.36 |
Rph3a |
rabphilin 3A |
262 |
0.91 |
| chr1_157243489_157244692 | 5.14 |
Rasal2 |
RAS protein activator like 2 |
400 |
0.88 |
| chr11_120046502_120047174 | 4.82 |
Aatk |
apoptosis-associated tyrosine kinase |
232 |
0.86 |
| chr3_26151491_26152156 | 4.63 |
Nlgn1 |
neuroligin 1 |
1484 |
0.58 |
| chr16_42336507_42337221 | 4.49 |
Gap43 |
growth associated protein 43 |
3787 |
0.3 |
| chr4_125489522_125489673 | 4.46 |
Grik3 |
glutamate receptor, ionotropic, kainate 3 |
1103 |
0.52 |
| chr9_73967774_73969221 | 4.44 |
Unc13c |
unc-13 homolog C |
469 |
0.88 |
| chr12_12938033_12939196 | 4.36 |
Mycn |
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
2002 |
0.23 |
| chr6_113501346_113502215 | 4.36 |
Prrt3 |
proline-rich transmembrane protein 3 |
38 |
0.94 |
| chr1_169746128_169746823 | 4.19 |
Rgs4 |
regulator of G-protein signaling 4 |
1148 |
0.55 |
| chr4_103619552_103620735 | 4.12 |
Dab1 |
disabled 1 |
478 |
0.8 |
| chr7_62420525_62420893 | 4.10 |
Mkrn3 |
makorin, ring finger protein, 3 |
570 |
0.69 |
| chr10_69705909_69707430 | 4.03 |
Ank3 |
ankyrin 3, epithelial |
191 |
0.97 |
| chr1_146494200_146495557 | 4.02 |
Gm29514 |
predicted gene 29514 |
73 |
0.56 |
| chr8_94269367_94270471 | 3.89 |
Nup93 |
nucleoporin 93 |
1313 |
0.31 |
| chr9_52148115_52149635 | 3.76 |
Zc3h12c |
zinc finger CCCH type containing 12C |
19236 |
0.18 |
| chr2_180026738_180027280 | 3.71 |
Lsm14b |
LSM family member 14B |
611 |
0.61 |
| chr1_42700819_42701404 | 3.66 |
Pou3f3 |
POU domain, class 3, transcription factor 3 |
5343 |
0.14 |
| chr10_64086822_64087463 | 3.60 |
Lrrtm3 |
leucine rich repeat transmembrane neuronal 3 |
3105 |
0.4 |
| chr16_41534559_41535426 | 3.55 |
Lsamp |
limbic system-associated membrane protein |
1573 |
0.55 |
| chr13_75941130_75942204 | 3.54 |
Rhobtb3 |
Rho-related BTB domain containing 3 |
2200 |
0.19 |
| chr15_25026077_25026585 | 3.46 |
Gm2824 |
predicted gene 2824 |
1006 |
0.65 |
| chr18_69349496_69350227 | 3.40 |
Tcf4 |
transcription factor 4 |
917 |
0.69 |
| chr1_172485277_172486996 | 3.31 |
Igsf9 |
immunoglobulin superfamily, member 9 |
3822 |
0.12 |
| chr7_127636875_127637879 | 3.25 |
Gm44760 |
predicted gene 44760 |
20857 |
0.07 |
| chr3_107041271_107042100 | 3.12 |
AI504432 |
expressed sequence AI504432 |
2181 |
0.26 |
| chr4_15880938_15882113 | 3.06 |
Calb1 |
calbindin 1 |
259 |
0.9 |
| chr18_13967357_13968027 | 3.04 |
Zfp521 |
zinc finger protein 521 |
4028 |
0.33 |
| chr18_64466262_64466884 | 3.00 |
Fech |
ferrochelatase |
2247 |
0.25 |
| chr1_143640264_143641520 | 2.98 |
B3galt2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
228 |
0.59 |
| chr11_102609159_102609359 | 2.96 |
Fzd2 |
frizzled class receptor 2 |
4863 |
0.11 |
| chr9_75683375_75684591 | 2.94 |
Scg3 |
secretogranin III |
8 |
0.97 |
| chr15_72545072_72545331 | 2.92 |
Kcnk9 |
potassium channel, subfamily K, member 9 |
1139 |
0.57 |
| chr12_3370750_3371679 | 2.87 |
Gm48511 |
predicted gene, 48511 |
2785 |
0.18 |
| chr3_68574662_68574918 | 2.85 |
Schip1 |
schwannomin interacting protein 1 |
2545 |
0.32 |
| chr10_40885530_40885990 | 2.82 |
Wasf1 |
WAS protein family, member 1 |
1933 |
0.32 |
| chr3_147083384_147083762 | 2.80 |
Gm29771 |
predicted gene, 29771 |
12611 |
0.27 |
| chr13_51568504_51570156 | 2.79 |
Shc3 |
src homology 2 domain-containing transforming protein C3 |
89 |
0.98 |
| chr18_31445092_31445474 | 2.79 |
Syt4 |
synaptotagmin IV |
2123 |
0.26 |
| chr3_34691662_34692725 | 2.70 |
Gm43206 |
predicted gene 43206 |
9560 |
0.13 |
| chr2_54082218_54082724 | 2.65 |
Rprm |
reprimo, TP53 dependent G2 arrest mediator candidate |
3081 |
0.32 |
| chr7_79498955_79500626 | 2.62 |
Mir9-3hg |
Mir9-3 host gene |
236 |
0.84 |
| chr11_84011128_84011766 | 2.61 |
Synrg |
synergin, gamma |
7754 |
0.2 |
| chr1_59515761_59516856 | 2.57 |
Gm973 |
predicted gene 973 |
44 |
0.96 |
| chr5_100120519_100121423 | 2.57 |
Tmem150c |
transmembrane protein 150C |
1825 |
0.31 |
| chrX_72655248_72656218 | 2.56 |
Gabra3 |
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
453 |
0.82 |
| chr5_124351883_124352230 | 2.52 |
Cdk2ap1 |
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
177 |
0.91 |
| chr5_77058285_77058946 | 2.51 |
Thegl |
theg spermatid protein like |
21561 |
0.11 |
| chr11_64956791_64957700 | 2.48 |
Elac2 |
elaC ribonuclease Z 2 |
21793 |
0.21 |
| chr6_94668134_94668755 | 2.48 |
Lrig1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
10421 |
0.23 |
| chrX_110814107_110814258 | 2.47 |
Pou3f4 |
POU domain, class 3, transcription factor 4 |
98 |
0.97 |
| chr14_55853912_55855354 | 2.41 |
Nynrin |
NYN domain and retroviral integrase containing |
597 |
0.55 |
| chr1_159523756_159524611 | 2.37 |
Tnr |
tenascin R |
342 |
0.91 |
| chr8_45908980_45909131 | 2.36 |
Pdlim3 |
PDZ and LIM domain 3 |
5977 |
0.13 |
| chrX_101299067_101300498 | 2.35 |
Nlgn3 |
neuroligin 3 |
572 |
0.58 |
| chr14_63609465_63609960 | 2.34 |
Gm15918 |
predicted gene 15918 |
2340 |
0.26 |
| chr9_53974078_53975628 | 2.27 |
Elmod1 |
ELMO/CED-12 domain containing 1 |
212 |
0.95 |
| chr8_33747278_33748028 | 2.27 |
Smim18 |
small integral membrane protein 18 |
117 |
0.95 |
| chr9_108824114_108825614 | 2.26 |
Gm35025 |
predicted gene, 35025 |
3 |
0.87 |
| chr8_94869714_94871671 | 2.26 |
Dok4 |
docking protein 4 |
512 |
0.65 |
| chr5_112520948_112521383 | 2.14 |
Sez6l |
seizure related 6 homolog like |
46038 |
0.1 |
| chr7_82056702_82057245 | 2.13 |
Gm20744 |
predicted gene, 20744 |
13414 |
0.15 |
| chr6_31088290_31089465 | 2.13 |
Lncpint |
long non-protein coding RNA, Trp53 induced transcript |
406 |
0.76 |
| chr12_53246370_53247488 | 2.11 |
Npas3 |
neuronal PAS domain protein 3 |
1228 |
0.61 |
| chr7_44335441_44336039 | 2.10 |
Shank1 |
SH3 and multiple ankyrin repeat domains 1 |
276 |
0.76 |
| chrX_6777615_6778651 | 2.09 |
Dgkk |
diacylglycerol kinase kappa |
1173 |
0.64 |
| chr19_44723625_44724799 | 2.08 |
Gm35610 |
predicted gene, 35610 |
15667 |
0.14 |
| chr11_117657020_117658010 | 2.08 |
Tnrc6c |
trinucleotide repeat containing 6C |
2702 |
0.26 |
| chr11_4948719_4949803 | 2.07 |
Nefh |
neurofilament, heavy polypeptide |
1197 |
0.38 |
| chr1_135510541_135510981 | 2.05 |
Nav1 |
neuron navigator 1 |
310 |
0.87 |
| chr13_83984481_83985348 | 2.04 |
Gm4241 |
predicted gene 4241 |
3077 |
0.26 |
| chr5_148391236_148391521 | 2.02 |
Slc7a1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
1437 |
0.49 |
| chr17_30805973_30806124 | 2.02 |
Dnah8 |
dynein, axonemal, heavy chain 8 |
2129 |
0.25 |
| chr13_81945552_81946341 | 2.00 |
F830210D05Rik |
RIKEN cDNA F830210D05 gene |
16259 |
0.25 |
| chr8_94998841_94999505 | 1.99 |
Adgrg1 |
adhesion G protein-coupled receptor G1 |
1708 |
0.25 |
| chr14_84452238_84452679 | 1.99 |
Pcdh17 |
protocadherin 17 |
3951 |
0.28 |
| chr3_5224377_5225076 | 1.97 |
Zfhx4 |
zinc finger homeodomain 4 |
3221 |
0.24 |
| chr17_56472644_56474083 | 1.96 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
1264 |
0.39 |
| chr5_141242772_141243546 | 1.96 |
Sdk1 |
sidekick cell adhesion molecule 1 |
1241 |
0.47 |
| chr4_130794177_130795043 | 1.95 |
Sdc3 |
syndecan 3 |
1906 |
0.21 |
| chr1_194646001_194646228 | 1.91 |
Plxna2 |
plexin A2 |
1258 |
0.41 |
| chr4_82499658_82501360 | 1.91 |
Nfib |
nuclear factor I/B |
1193 |
0.5 |
| chr7_138940266_138941503 | 1.91 |
Jakmip3 |
janus kinase and microtubule interacting protein 3 |
154 |
0.93 |
| chr6_147261502_147261714 | 1.89 |
Pthlh |
parathyroid hormone-like peptide |
2559 |
0.25 |
| chr19_55898749_55899177 | 1.87 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
654 |
0.79 |
| chr16_77662281_77662497 | 1.86 |
9530003O04Rik |
RIKEN cDNA 9530003O04 gene |
7858 |
0.11 |
| chr6_114133449_114133914 | 1.85 |
Slc6a11 |
solute carrier family 6 (neurotransmitter transporter, GABA), member 11 |
2262 |
0.31 |
| chr3_89338092_89338353 | 1.85 |
Efna4 |
ephrin A4 |
194 |
0.63 |
| chr18_79336296_79336940 | 1.84 |
Gm20593 |
predicted gene, 20593 |
11816 |
0.24 |
| chr1_137899773_137901229 | 1.81 |
Gm4258 |
predicted gene 4258 |
1903 |
0.18 |
| chr4_129496518_129496993 | 1.78 |
Fam229a |
family with sequence similarity 229, member A |
5515 |
0.09 |
| chr8_12324957_12325822 | 1.78 |
Gm33175 |
predicted gene, 33175 |
5909 |
0.18 |
| chr19_53340390_53340659 | 1.76 |
Mxi1 |
MAX interactor 1, dimerization protein |
10116 |
0.13 |
| chr2_70558567_70559918 | 1.75 |
Gad1 |
glutamate decarboxylase 1 |
2800 |
0.19 |
| chr5_37245879_37246957 | 1.74 |
Crmp1 |
collapsin response mediator protein 1 |
573 |
0.76 |
| chr10_4709783_4710728 | 1.70 |
Esr1 |
estrogen receptor 1 (alpha) |
100 |
0.98 |
| chr13_54744689_54744840 | 1.70 |
Gprin1 |
G protein-regulated inducer of neurite outgrowth 1 |
4905 |
0.13 |
| chr7_87246018_87247310 | 1.69 |
Nox4 |
NADPH oxidase 4 |
15 |
0.98 |
| chrX_7728575_7729385 | 1.67 |
Praf2 |
PRA1 domain family 2 |
541 |
0.49 |
| chr3_80798726_80799058 | 1.67 |
Gria2 |
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
3687 |
0.31 |
| chr5_138993920_138994071 | 1.66 |
Pdgfa |
platelet derived growth factor, alpha |
286 |
0.84 |
| chr2_19414639_19415225 | 1.64 |
Gm33940 |
predicted gene, 33940 |
13114 |
0.13 |
| chr6_6870101_6870700 | 1.64 |
Dlx6os1 |
distal-less homeobox 6, opposite strand 1 |
1192 |
0.35 |
| chr7_16919610_16920549 | 1.62 |
Calm3 |
calmodulin 3 |
2876 |
0.12 |
| chr1_43005796_43006609 | 1.62 |
Gpr45 |
G protein-coupled receptor 45 |
25858 |
0.14 |
| chr5_25472059_25472730 | 1.61 |
Gm18343 |
predicted gene, 18343 |
22089 |
0.11 |
| chr5_131631448_131631721 | 1.59 |
Gm42591 |
predicted gene 42591 |
6043 |
0.12 |
| chr5_113901213_113901683 | 1.58 |
Gm43336 |
predicted gene 43336 |
3450 |
0.14 |
| chr4_98108018_98108324 | 1.58 |
Gm12691 |
predicted gene 12691 |
38428 |
0.2 |
| chr19_55893416_55893964 | 1.56 |
Tcf7l2 |
transcription factor 7 like 2, T cell specific, HMG box |
338 |
0.92 |
| chr3_96605760_96605911 | 1.56 |
6330549D23Rik |
RIKEN cDNA 6330549D23 gene |
2890 |
0.09 |
| chr11_120259845_120260692 | 1.55 |
Gm47297 |
predicted gene, 47297 |
14219 |
0.1 |
| chr5_53707130_53707688 | 1.55 |
Cckar |
cholecystokinin A receptor |
151 |
0.97 |
| chr1_182410542_182411314 | 1.55 |
Trp53bp2 |
transformation related protein 53 binding protein 2 |
1756 |
0.25 |
| chr19_29044756_29045015 | 1.53 |
1700018L02Rik |
RIKEN cDNA 1700018L02 gene |
1360 |
0.29 |
| chr7_127615293_127615738 | 1.53 |
Zfp629 |
zinc finger protein 629 |
243 |
0.81 |
| chr7_27445809_27447233 | 1.50 |
Sptbn4 |
spectrin beta, non-erythrocytic 4 |
4 |
0.94 |
| chr6_28832419_28832731 | 1.50 |
Lrrc4 |
leucine rich repeat containing 4 |
828 |
0.63 |
| chr2_117141382_117141533 | 1.49 |
Spred1 |
sprouty protein with EVH-1 domain 1, related sequence |
19819 |
0.19 |
| chr5_149331898_149332049 | 1.49 |
BC028471 |
cDNA sequence BC028471 |
35663 |
0.08 |
| chr3_34197711_34199105 | 1.48 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
1544 |
0.41 |
| chr1_132199896_132201879 | 1.48 |
Lemd1 |
LEM domain containing 1 |
76 |
0.95 |
| chr1_79759313_79760206 | 1.47 |
Wdfy1 |
WD repeat and FYVE domain containing 1 |
1962 |
0.28 |
| chr6_58904659_58904810 | 1.46 |
Nap1l5 |
nucleosome assembly protein 1-like 5 |
1939 |
0.28 |
| chr8_90955427_90955995 | 1.45 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
276 |
0.9 |
| chr17_93201490_93204144 | 1.45 |
Adcyap1 |
adenylate cyclase activating polypeptide 1 |
741 |
0.65 |
| chr5_111054948_111055658 | 1.42 |
Gm42780 |
predicted gene 42780 |
26751 |
0.15 |
| chr7_105776779_105776930 | 1.42 |
Dchs1 |
dachsous cadherin related 1 |
10698 |
0.09 |
| chr4_35844509_35845617 | 1.42 |
Lingo2 |
leucine rich repeat and Ig domain containing 2 |
141 |
0.98 |
| chr10_119691568_119692137 | 1.41 |
Grip1 |
glutamate receptor interacting protein 1 |
202 |
0.95 |
| chr5_52475943_52477197 | 1.40 |
Ccdc149 |
coiled-coil domain containing 149 |
5049 |
0.19 |
| chr12_36315313_36316308 | 1.40 |
Sostdc1 |
sclerostin domain containing 1 |
1671 |
0.31 |
| chr15_37457149_37457300 | 1.36 |
Ncald |
neurocalcin delta |
1317 |
0.36 |
| chr5_51991017_51991506 | 1.36 |
Gm43175 |
predicted gene 43175 |
80330 |
0.07 |
| chr3_129532189_129532340 | 1.36 |
Elovl6 |
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
91 |
0.54 |
| chr1_86302981_86304185 | 1.35 |
B3gnt7 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
52 |
0.86 |
| chr1_72875853_72876281 | 1.35 |
Igfbp5 |
insulin-like growth factor binding protein 5 |
1183 |
0.53 |
| chrX_144153485_144154716 | 1.35 |
A730046J19Rik |
RIKEN cDNA A730046J19 gene |
405 |
0.85 |
| chr3_97763340_97765161 | 1.34 |
Pde4dip |
phosphodiesterase 4D interacting protein (myomegalin) |
3681 |
0.23 |
| chr6_85501969_85503009 | 1.34 |
Fbxo41 |
F-box protein 41 |
369 |
0.8 |
| chr2_79637031_79637387 | 1.34 |
Itprid2 |
ITPR interacting domain containing 2 |
1748 |
0.48 |
| chr3_34314559_34314742 | 1.33 |
Gm38505 |
predicted gene, 38505 |
37062 |
0.17 |
| chr18_38596322_38597743 | 1.32 |
Spry4 |
sprouty RTK signaling antagonist 4 |
4383 |
0.18 |
| chr14_93887180_93887734 | 1.32 |
Pcdh9 |
protocadherin 9 |
1233 |
0.6 |
| chr2_30443959_30444632 | 1.30 |
Ptpa |
protein phosphatase 2 protein activator |
1076 |
0.39 |
| chr9_41952303_41952488 | 1.30 |
Sorl1 |
sortilin-related receptor, LDLR class A repeats-containing |
22192 |
0.17 |
| chr2_159002562_159003405 | 1.30 |
Gm44319 |
predicted gene, 44319 |
66880 |
0.12 |
| chr10_96825969_96826706 | 1.29 |
Gm48521 |
predicted gene, 48521 |
25298 |
0.15 |
| chr7_44383854_44385056 | 1.29 |
Syt3 |
synaptotagmin III |
149 |
0.85 |
| chr3_89232897_89233538 | 1.28 |
Muc1 |
mucin 1, transmembrane |
1746 |
0.12 |
| chr11_102061379_102062137 | 1.28 |
Cd300lg |
CD300 molecule like family member G |
20249 |
0.08 |
| chrX_64273727_64274331 | 1.26 |
Slitrk4 |
SLIT and NTRK-like family, member 4 |
1793 |
0.49 |
| chr17_47920777_47921128 | 1.26 |
Gm15556 |
predicted gene 15556 |
1426 |
0.31 |
| chr5_131596575_131596859 | 1.26 |
Gm42589 |
predicted gene 42589 |
8687 |
0.12 |
| chr5_31494383_31494599 | 1.25 |
Gpn1 |
GPN-loop GTPase 1 |
250 |
0.73 |
| chr11_97808223_97808514 | 1.24 |
Lasp1 |
LIM and SH3 protein 1 |
1866 |
0.17 |
| chr8_12386921_12387188 | 1.24 |
Sox1ot |
Sox1 overlapping transcript |
1283 |
0.34 |
| chr11_77933689_77934630 | 1.22 |
Sez6 |
seizure related gene 6 |
3120 |
0.17 |
| chr15_99690869_99691020 | 1.22 |
Asic1 |
acid-sensing (proton-gated) ion channel 1 |
1289 |
0.23 |
| chr7_4690514_4690708 | 1.21 |
Brsk1 |
BR serine/threonine kinase 1 |
7 |
0.94 |
| chr1_177562035_177562186 | 1.20 |
Gm37280 |
predicted gene, 37280 |
32412 |
0.16 |
| chr6_58903282_58903683 | 1.20 |
Herc3 |
hect domain and RLD 3 |
1734 |
0.32 |
| chr4_82507188_82507431 | 1.19 |
Gm11266 |
predicted gene 11266 |
707 |
0.62 |
| chr1_163574713_163575210 | 1.18 |
Gm29500 |
predicted gene 29500 |
32416 |
0.18 |
| chrX_158923078_158924649 | 1.17 |
Gm5764 |
predicted gene 5764 |
90428 |
0.09 |
| chr4_66402659_66403225 | 1.17 |
Astn2 |
astrotactin 2 |
1541 |
0.52 |
| chr14_32825387_32825835 | 1.15 |
Fam170b |
family with sequence similarity 170, member B |
8351 |
0.18 |
| chr7_45313085_45313236 | 1.13 |
Trpm4 |
transient receptor potential cation channel, subfamily M, member 4 |
1592 |
0.16 |
| chr1_66324716_66324867 | 1.12 |
Map2 |
microtubule-associated protein 2 |
2689 |
0.25 |
| chr11_97627602_97627753 | 1.12 |
Epop |
elongin BC and polycomb repressive complex 2 associated protein |
2025 |
0.18 |
| chr3_139860647_139861358 | 1.12 |
Gm43678 |
predicted gene 43678 |
48238 |
0.17 |
| chr11_28697846_28697997 | 1.10 |
2810471M01Rik |
RIKEN cDNA 2810471M01 gene |
16357 |
0.16 |
| chr15_98807369_98808449 | 1.10 |
Ddn |
dendrin |
16 |
0.92 |
| chr6_64733770_64735303 | 1.09 |
Atoh1 |
atonal bHLH transcription factor 1 |
5411 |
0.3 |
| chr6_28832773_28833300 | 1.09 |
Lrrc4 |
leucine rich repeat containing 4 |
1289 |
0.46 |
| chr12_88725972_88726370 | 1.09 |
Nrxn3 |
neurexin III |
490 |
0.84 |
| chr7_90386713_90387752 | 1.08 |
Sytl2 |
synaptotagmin-like 2 |
127 |
0.96 |
| chr2_26909958_26910571 | 1.08 |
Med22 |
mediator complex subunit 22 |
305 |
0.52 |
| chr6_135527026_135527177 | 1.07 |
Gm25136 |
predicted gene, 25136 |
57091 |
0.13 |
| chr10_7792967_7793511 | 1.06 |
Ppil4 |
peptidylprolyl isomerase (cyclophilin)-like 4 |
318 |
0.54 |
| chr2_16355730_16357176 | 1.06 |
Plxdc2 |
plexin domain containing 2 |
149 |
0.98 |
| chr7_30444318_30445408 | 1.06 |
Aplp1 |
amyloid beta (A4) precursor-like protein 1 |
267 |
0.76 |
| chr4_24897578_24897938 | 1.06 |
Ndufaf4 |
NADH:ubiquinone oxidoreductase complex assembly factor 4 |
325 |
0.9 |
| chr2_6873585_6874006 | 1.05 |
Celf2 |
CUGBP, Elav-like family member 2 |
1198 |
0.49 |
| chr16_11985754_11986188 | 1.04 |
Shisa9 |
shisa family member 9 |
1251 |
0.58 |
| chr5_115908055_115909691 | 1.04 |
Cit |
citron |
1403 |
0.37 |
| chr19_45717472_45718323 | 1.03 |
Fgf8 |
fibroblast growth factor 8 |
24787 |
0.12 |
| chr10_6979289_6980565 | 1.02 |
Ipcef1 |
interaction protein for cytohesin exchange factors 1 |
216 |
0.96 |
| chr6_116192607_116193328 | 1.02 |
Tmcc1 |
transmembrane and coiled coil domains 1 |
519 |
0.66 |
| chr3_130922899_130923995 | 1.01 |
Gm43349 |
predicted gene 43349 |
1390 |
0.29 |
| chr7_122670691_122671255 | 1.01 |
Cacng3 |
calcium channel, voltage-dependent, gamma subunit 3 |
418 |
0.85 |
| chr6_124996427_124996698 | 1.00 |
Pianp |
PILR alpha associated neural protein |
132 |
0.9 |
| chr7_119861356_119862025 | 1.00 |
Dcun1d3 |
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
114 |
0.95 |
| chr5_139537301_139537452 | 1.00 |
Uncx |
UNC homeobox |
6118 |
0.17 |
| chr16_44138294_44139384 | 1.00 |
Atp6v1a |
ATPase, H+ transporting, lysosomal V1 subunit A |
141 |
0.92 |
| chr4_82762782_82763099 | 0.99 |
Nfib |
nuclear factor I/B |
57190 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 1.1 | 3.2 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 1.0 | 3.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.9 | 2.6 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.8 | 4.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.7 | 2.2 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.7 | 1.4 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.6 | 2.5 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.5 | 1.6 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.5 | 3.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.5 | 2.8 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.5 | 1.4 | GO:2000969 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.4 | 1.3 | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.4 | 1.7 | GO:0046959 | habituation(GO:0046959) |
| 0.3 | 1.4 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.3 | 0.7 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.3 | 0.9 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.3 | 0.9 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 0.9 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 1.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 | 0.3 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.3 | 0.5 | GO:0072240 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.3 | 0.8 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.3 | 0.8 | GO:1903935 | response to sodium arsenite(GO:1903935) |
| 0.3 | 0.8 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.3 | 0.8 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.2 | 6.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.5 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.2 | 0.7 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 0.7 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.2 | 2.8 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 1.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 2.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 1.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.6 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.2 | 1.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.2 | 0.4 | GO:0072144 | glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.2 | 3.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 0.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 1.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 0.5 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.2 | 1.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.2 | 0.5 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.2 | 1.5 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.2 | 1.3 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.2 | 0.5 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.2 | 0.6 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.2 | 1.7 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.2 | 0.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 0.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.4 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 1.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.4 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.1 | 0.8 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.7 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.1 | 0.4 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 1.3 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 0.9 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 1.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 1.0 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.3 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.3 | GO:0009629 | response to gravity(GO:0009629) |
| 0.1 | 0.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.3 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.1 | 1.3 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 1.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.3 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.1 | 0.8 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 4.1 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 | 0.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.7 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 2.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.1 | GO:0061325 | cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.1 | 0.3 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.3 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.6 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 1.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 3.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.0 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.5 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.7 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.2 | GO:0098910 | regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.1 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 2.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.4 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.5 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.1 | 0.2 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 | 0.1 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.1 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.2 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.1 | 1.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.5 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.1 | 0.2 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.2 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.7 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 0.2 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.1 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.1 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.1 | 0.2 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 | 0.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.1 | GO:0021648 | vestibulocochlear nerve morphogenesis(GO:0021648) |
| 0.1 | 1.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.1 | GO:1902966 | protein localization to early endosome(GO:1902946) regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.0 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 1.0 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.4 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.0 | GO:2000054 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.5 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0033505 | floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.2 | GO:0060696 | regulation of phospholipid catabolic process(GO:0060696) |
| 0.0 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 1.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.2 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.1 | GO:0045472 | response to ether(GO:0045472) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 1.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 2.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.0 | GO:0036515 | dopaminergic neuron axon guidance(GO:0036514) serotonergic neuron axon guidance(GO:0036515) planar cell polarity pathway involved in axon guidance(GO:1904938) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.4 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.3 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 1.0 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.0 | GO:1902837 | amino acid import into cell(GO:1902837) L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.3 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.8 | GO:0090662 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.5 | GO:0060998 | regulation of dendritic spine development(GO:0060998) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.0 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.3 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.0 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.0 | 1.8 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.0 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.0 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.1 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.0 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.1 | GO:0009068 | aspartate family amino acid catabolic process(GO:0009068) |
| 0.0 | 0.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.0 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.1 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.0 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.2 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.0 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.0 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.1 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.0 | 0.1 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.2 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.5 | GO:2001259 | positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.6 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.0 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.0 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.0 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.0 | 0.0 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.0 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.0 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 0.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.0 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.0 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.1 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.0 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.0 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.0 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.0 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.1 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.4 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.0 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.3 | GO:0008542 | visual learning(GO:0008542) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.7 | 2.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.4 | 4.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.4 | 4.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.4 | 1.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 4.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 0.8 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.2 | 0.4 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.2 | 1.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.5 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 0.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 4.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 3.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 1.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 0.8 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 5.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.1 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.5 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.4 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 5.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 1.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 1.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 2.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 6.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.2 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.0 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 1.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 2.9 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 5.2 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.0 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 1.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.0 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.0 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.4 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.0 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.0 | GO:0061574 | ASAP complex(GO:0061574) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.6 | 2.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.6 | 1.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.6 | 3.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.5 | 2.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.4 | 1.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.4 | 4.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 3.5 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.4 | 2.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 3.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 1.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.3 | 1.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.3 | 1.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.3 | 1.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 0.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 1.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 3.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 0.7 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.2 | 0.7 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.2 | 0.6 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.2 | 0.8 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.2 | 0.7 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 2.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 0.5 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 2.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.2 | 0.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 0.5 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.1 | 0.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 3.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.7 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 2.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.8 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 3.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.8 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.5 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 11.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0044606 | thiamine-pyrophosphatase activity(GO:0004787) UDP-2,3-diacylglucosamine hydrolase activity(GO:0008758) dATP pyrophosphohydrolase activity(GO:0008828) dihydroneopterin monophosphate phosphatase activity(GO:0019176) dihydroneopterin triphosphate pyrophosphohydrolase activity(GO:0019177) dTTP diphosphatase activity(GO:0036218) phosphocholine hydrolase activity(GO:0044606) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.2 | GO:0016934 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.2 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0070635 | purine-specific mismatch base pair DNA N-glycosylase activity(GO:0000701) single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.9 | GO:0080062 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.3 | GO:0052713 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 1.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.5 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:1990190 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.4 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.6 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 2.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 6.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 3.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 4.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 3.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 0.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.2 | 2.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.6 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 1.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 0.9 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 1.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |