Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
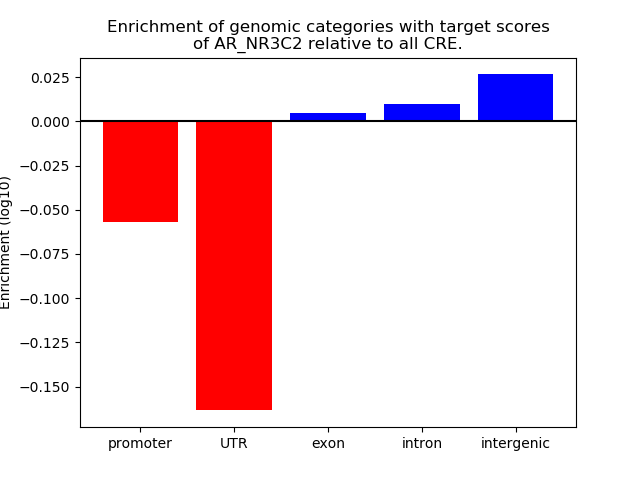
Results for AR_NR3C2
Z-value: 1.48
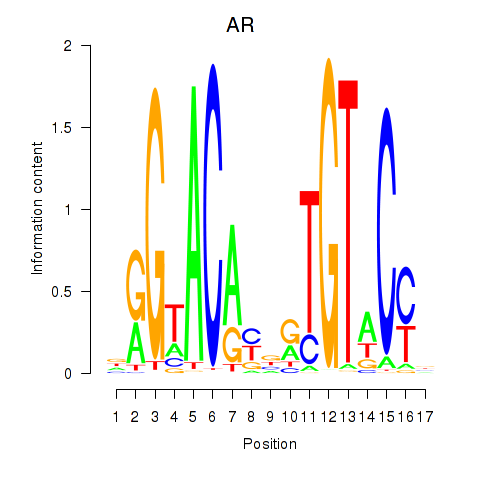
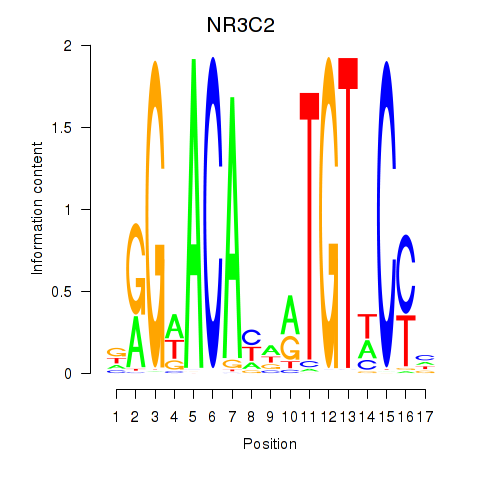
Transcription factors associated with AR_NR3C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AR
|
ENSG00000169083.11 | androgen receptor |
|
NR3C2
|
ENSG00000151623.10 | nuclear receptor subfamily 3 group C member 2 |
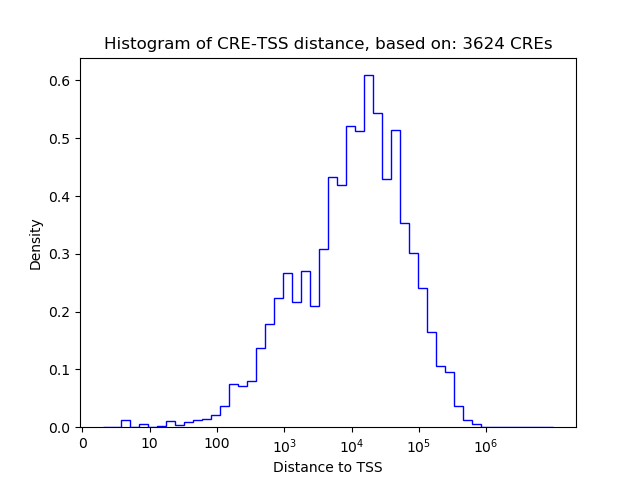
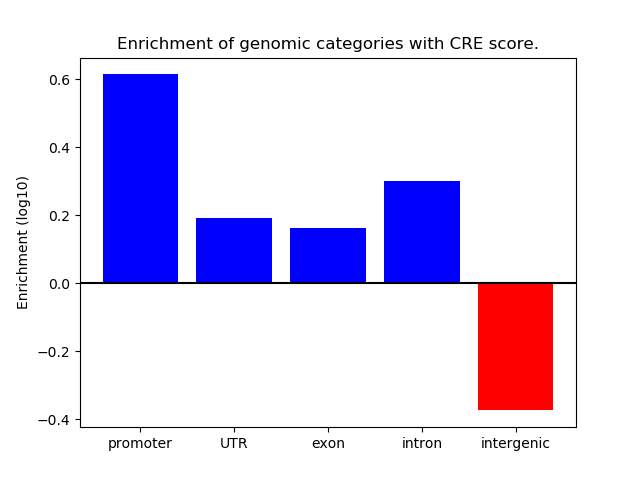
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_66915365_66915591 | AR | 126747 | 0.062179 | 0.86 | 2.7e-03 | Click! |
| chrX_66946272_66946453 | AR | 157631 | 0.042980 | -0.75 | 2.1e-02 | Click! |
| chrX_66955172_66955323 | AR | 166516 | 0.039019 | -0.66 | 5.5e-02 | Click! |
| chrX_67004998_67005149 | AR | 216342 | 0.024177 | -0.65 | 5.6e-02 | Click! |
| chrX_67000391_67000546 | AR | 211737 | 0.025171 | 0.62 | 7.5e-02 | Click! |
| chr4_149276556_149276707 | NR3C2 | 81381 | 0.112390 | -0.82 | 6.3e-03 | Click! |
| chr4_149229242_149229393 | NR3C2 | 128695 | 0.056491 | -0.79 | 1.2e-02 | Click! |
| chr4_149350438_149350589 | NR3C2 | 7499 | 0.323590 | -0.77 | 1.5e-02 | Click! |
| chr4_149353611_149353762 | NR3C2 | 4326 | 0.357265 | -0.73 | 2.7e-02 | Click! |
| chr4_149298361_149298512 | NR3C2 | 59576 | 0.160340 | -0.71 | 3.1e-02 | Click! |
Activity of the AR_NR3C2 motif across conditions
Conditions sorted by the z-value of the AR_NR3C2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
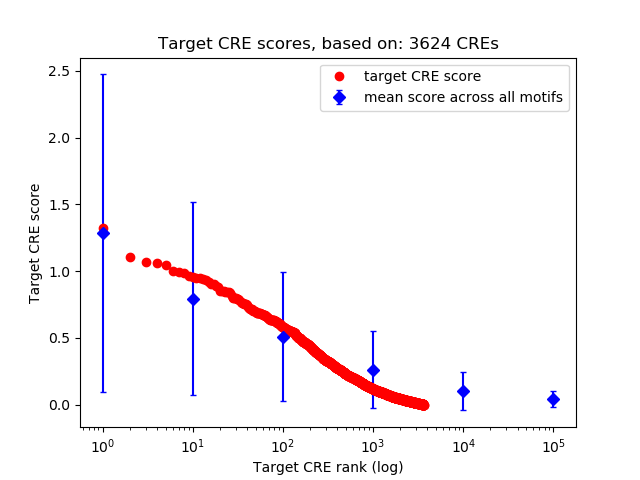
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_100004430_100004581 | 1.32 |
CCDC85C |
coiled-coil domain containing 85C |
2080 |
0.29 |
| chr2_19848269_19848420 | 1.10 |
TTC32 |
tetratricopeptide repeat domain 32 |
253058 |
0.02 |
| chr1_66932158_66932309 | 1.07 |
SGIP1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
66833 |
0.12 |
| chr18_34021656_34021807 | 1.06 |
FHOD3 |
formin homology 2 domain containing 3 |
102783 |
0.08 |
| chr15_63045655_63045939 | 1.05 |
TLN2 |
talin 2 |
4992 |
0.22 |
| chr2_3474011_3474213 | 1.00 |
TRAPPC12 |
trafficking protein particle complex 12 |
4668 |
0.18 |
| chr6_141179242_141179445 | 0.99 |
ENSG00000264390 |
. |
174392 |
0.04 |
| chr12_27900681_27900869 | 0.99 |
ENSG00000252585 |
. |
5032 |
0.13 |
| chr10_30236708_30236859 | 0.96 |
KIAA1462 |
KIAA1462 |
111670 |
0.07 |
| chr1_22196645_22196796 | 0.95 |
HSPG2 |
heparan sulfate proteoglycan 2 |
4479 |
0.22 |
| chr2_48710705_48710856 | 0.95 |
ENSG00000202227 |
. |
18383 |
0.18 |
| chr20_34685745_34686271 | 0.95 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
5249 |
0.2 |
| chr10_30235923_30236074 | 0.94 |
KIAA1462 |
KIAA1462 |
112455 |
0.07 |
| chr11_12434149_12434300 | 0.94 |
PARVA |
parvin, alpha |
14436 |
0.24 |
| chr1_244512198_244512349 | 0.92 |
C1orf100 |
chromosome 1 open reading frame 100 |
3664 |
0.3 |
| chr4_184558613_184558764 | 0.90 |
ENSG00000252157 |
. |
16141 |
0.14 |
| chr20_56592771_56592922 | 0.90 |
ENSG00000221451 |
. |
41772 |
0.15 |
| chr3_48731676_48731827 | 0.89 |
IP6K2 |
inositol hexakisphosphate kinase 2 |
1264 |
0.31 |
| chr2_145769885_145770036 | 0.88 |
ENSG00000253036 |
. |
322678 |
0.01 |
| chr19_3336838_3336989 | 0.85 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
22648 |
0.15 |
| chr8_102458007_102458158 | 0.85 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
46578 |
0.13 |
| chr16_57687826_57687988 | 0.85 |
GPR56 |
G protein-coupled receptor 56 |
764 |
0.56 |
| chr1_180827525_180827764 | 0.85 |
ENSG00000266825 |
. |
30516 |
0.17 |
| chr4_57594846_57594997 | 0.85 |
HOPX |
HOP homeobox |
46856 |
0.13 |
| chr18_45856611_45856862 | 0.84 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
8026 |
0.26 |
| chr2_47063884_47064035 | 0.83 |
AC016722.3 |
|
16676 |
0.14 |
| chr1_109371861_109372012 | 0.82 |
AKNAD1 |
AKNA domain containing 1 |
23403 |
0.15 |
| chr6_161724528_161724679 | 0.80 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
29510 |
0.26 |
| chr3_53184032_53184571 | 0.80 |
PRKCD |
protein kinase C, delta |
5724 |
0.19 |
| chr1_167766173_167766370 | 0.80 |
RP1-313L4.3 |
|
23373 |
0.19 |
| chr20_4151764_4151915 | 0.79 |
SMOX |
spermine oxidase |
518 |
0.81 |
| chr17_46561232_46561443 | 0.79 |
ENSG00000206805 |
. |
4878 |
0.14 |
| chr20_45543740_45543891 | 0.78 |
EYA2 |
eyes absent homolog 2 (Drosophila) |
20552 |
0.22 |
| chr3_40158038_40158189 | 0.77 |
MYRIP |
myosin VIIA and Rab interacting protein |
16611 |
0.25 |
| chr17_43170864_43171027 | 0.76 |
PLCD3 |
phospholipase C, delta 3 |
19971 |
0.1 |
| chr2_241190811_241190962 | 0.76 |
ENSG00000221412 |
. |
10391 |
0.24 |
| chr7_143209652_143209944 | 0.76 |
TAS2R41 |
taste receptor, type 2, member 41 |
34832 |
0.12 |
| chr14_93439391_93439542 | 0.76 |
CHGA |
chromogranin A (parathyroid secretory protein 1) |
49945 |
0.14 |
| chr1_223888318_223888469 | 0.76 |
CAPN2 |
calpain 2, (m/II) large subunit |
902 |
0.61 |
| chr10_74877735_74877886 | 0.75 |
RP11-152N13.16 |
|
7353 |
0.1 |
| chr12_13331696_13332290 | 0.73 |
EMP1 |
epithelial membrane protein 1 |
17657 |
0.23 |
| chr3_99640879_99641030 | 0.72 |
ENSG00000264897 |
. |
42288 |
0.16 |
| chr2_33383859_33384010 | 0.72 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
24210 |
0.25 |
| chr4_24913711_24913879 | 0.72 |
CCDC149 |
coiled-coil domain containing 149 |
713 |
0.79 |
| chr1_11219246_11219397 | 0.71 |
ENSG00000253086 |
. |
6914 |
0.12 |
| chr6_86173389_86173540 | 0.71 |
NT5E |
5'-nucleotidase, ecto (CD73) |
7635 |
0.28 |
| chr2_28021942_28022093 | 0.70 |
AC110084.1 |
|
12831 |
0.16 |
| chr8_56861171_56861322 | 0.70 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
8453 |
0.16 |
| chr8_124562734_124562885 | 0.70 |
FBXO32 |
F-box protein 32 |
9363 |
0.2 |
| chr10_13881950_13882101 | 0.69 |
FRMD4A |
FERM domain containing 4A |
18887 |
0.22 |
| chr2_238787700_238787851 | 0.69 |
ENSG00000263723 |
. |
9228 |
0.2 |
| chr17_15866796_15866947 | 0.69 |
ADORA2B |
adenosine A2b receptor |
18640 |
0.15 |
| chr5_150159787_150160102 | 0.69 |
AC010441.1 |
|
2077 |
0.24 |
| chr6_155593510_155593784 | 0.69 |
CLDN20 |
claudin 20 |
8500 |
0.2 |
| chr14_55925449_55925600 | 0.69 |
TBPL2 |
TATA box binding protein like 2 |
2080 |
0.38 |
| chr9_3863683_3863834 | 0.68 |
RP11-252M18.3 |
|
11826 |
0.25 |
| chr6_151357426_151357577 | 0.68 |
MTHFD1L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
609 |
0.72 |
| chr12_26441077_26441474 | 0.68 |
RP11-283G6.5 |
|
16422 |
0.18 |
| chr21_39612976_39613250 | 0.68 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
15550 |
0.24 |
| chr13_110161938_110162089 | 0.67 |
LINC00676 |
long intergenic non-protein coding RNA 676 |
218616 |
0.02 |
| chr4_170129389_170129540 | 0.67 |
RP11-327O17.2 |
|
6519 |
0.28 |
| chr9_130447197_130447348 | 0.67 |
ENSG00000264329 |
. |
5802 |
0.13 |
| chr4_169717957_169718108 | 0.67 |
PALLD |
palladin, cytoskeletal associated protein |
35124 |
0.16 |
| chr6_43636660_43636811 | 0.66 |
MRPS18A |
mitochondrial ribosomal protein S18A |
18793 |
0.12 |
| chr7_75678596_75678833 | 0.66 |
MDH2 |
malate dehydrogenase 2, NAD (mitochondrial) |
1235 |
0.32 |
| chr5_149878333_149878560 | 0.65 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
957 |
0.56 |
| chr4_41062724_41062875 | 0.65 |
ENSG00000199790 |
. |
23825 |
0.18 |
| chr4_6993000_6993535 | 0.65 |
AC097382.5 |
|
3801 |
0.14 |
| chr1_201886871_201887022 | 0.65 |
RP11-307B6.3 |
|
19718 |
0.1 |
| chr11_95254135_95254332 | 0.64 |
ENSG00000201204 |
. |
47695 |
0.19 |
| chr5_131604535_131604686 | 0.64 |
PDLIM4 |
PDZ and LIM domain 4 |
7468 |
0.16 |
| chr3_156534966_156535117 | 0.64 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
9048 |
0.29 |
| chr16_50237563_50237763 | 0.64 |
ADCY7 |
adenylate cyclase 7 |
42385 |
0.12 |
| chr5_95674311_95674759 | 0.64 |
PCSK1 |
proprotein convertase subtilisin/kexin type 1 |
93329 |
0.08 |
| chr10_34873395_34873546 | 0.64 |
PARD3 |
par-3 family cell polarity regulator |
157811 |
0.04 |
| chr13_109609770_109610065 | 0.64 |
MYO16 |
myosin XVI |
71400 |
0.13 |
| chr5_95401493_95401712 | 0.63 |
ENSG00000207578 |
. |
13240 |
0.25 |
| chr3_73085736_73086140 | 0.63 |
ENSG00000238959 |
. |
8979 |
0.16 |
| chr16_9142329_9142480 | 0.63 |
C16orf72 |
chromosome 16 open reading frame 72 |
43101 |
0.12 |
| chr22_24980060_24980211 | 0.63 |
GGT1 |
gamma-glutamyltransferase 1 |
417 |
0.78 |
| chr12_3309876_3310419 | 0.63 |
TSPAN9 |
tetraspanin 9 |
194 |
0.96 |
| chr17_79039549_79039700 | 0.63 |
BAIAP2 |
BAI1-associated protein 2 |
8090 |
0.13 |
| chr5_157810174_157810325 | 0.63 |
ENSG00000222626 |
. |
406285 |
0.01 |
| chr21_36696632_36696783 | 0.62 |
RUNX1 |
runt-related transcription factor 1 |
275066 |
0.01 |
| chr15_42694918_42695069 | 0.62 |
CAPN3 |
calpain 3, (p94) |
4 |
0.97 |
| chr18_46260220_46260632 | 0.62 |
RP11-426J5.2 |
|
45636 |
0.16 |
| chr21_30518493_30518644 | 0.62 |
ENSG00000201984 |
. |
6863 |
0.14 |
| chr2_102680238_102680787 | 0.61 |
IL1R1 |
interleukin 1 receptor, type I |
492 |
0.85 |
| chr22_50969649_50969871 | 0.61 |
ODF3B |
outer dense fiber of sperm tails 3B |
746 |
0.33 |
| chr5_119789870_119790021 | 0.61 |
PRR16 |
proline rich 16 |
10028 |
0.31 |
| chr9_15735715_15735866 | 0.61 |
CCDC171 |
coiled-coil domain containing 171 |
8713 |
0.26 |
| chr2_15310055_15310206 | 0.60 |
NBAS |
neuroblastoma amplified sequence |
20391 |
0.29 |
| chr1_198202205_198202356 | 0.60 |
NEK7 |
NIMA-related kinase 7 |
12351 |
0.31 |
| chr6_15504791_15504942 | 0.60 |
DTNBP1 |
dystrobrevin binding protein 1 |
43727 |
0.19 |
| chr13_94962334_94962485 | 0.60 |
GPC6-AS1 |
GPC6 antisense RNA 1 |
122164 |
0.06 |
| chr4_145621362_145621513 | 0.59 |
HHIP-AS1 |
HHIP antisense RNA 1 |
38928 |
0.19 |
| chr2_46542548_46542756 | 0.59 |
EPAS1 |
endothelial PAS domain protein 1 |
18111 |
0.25 |
| chr2_36681200_36681351 | 0.59 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
59558 |
0.12 |
| chr12_46887776_46887927 | 0.58 |
SLC38A2 |
solute carrier family 38, member 2 |
121201 |
0.06 |
| chr13_30700064_30700215 | 0.58 |
ENSG00000266816 |
. |
80697 |
0.11 |
| chr8_13282364_13282515 | 0.58 |
DLC1 |
deleted in liver cancer 1 |
74311 |
0.1 |
| chr8_96812845_96812996 | 0.58 |
ENSG00000223297 |
. |
121777 |
0.06 |
| chr4_86954278_86954588 | 0.57 |
RP13-514E23.1 |
|
19406 |
0.19 |
| chr22_27501498_27501649 | 0.57 |
ENSG00000200443 |
. |
69723 |
0.14 |
| chr8_67472226_67472377 | 0.57 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
52841 |
0.1 |
| chr22_47426182_47426436 | 0.57 |
ENSG00000221672 |
. |
182506 |
0.03 |
| chr8_120126669_120126820 | 0.57 |
RP11-278I4.2 |
|
45723 |
0.15 |
| chr1_205335200_205335351 | 0.57 |
LEMD1-AS1 |
LEMD1 antisense RNA 1 |
7105 |
0.17 |
| chr7_40261588_40261739 | 0.57 |
SUGCT |
succinylCoA:glutarate-CoA transferase |
87044 |
0.09 |
| chr6_155850479_155850743 | 0.57 |
NOX3 |
NADPH oxidase 3 |
73574 |
0.12 |
| chr1_183117616_183117767 | 0.56 |
RP11-181K3.4 |
|
7274 |
0.22 |
| chr12_53443486_53443637 | 0.56 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
119 |
0.93 |
| chr2_232539358_232539509 | 0.56 |
ENSG00000239202 |
. |
28449 |
0.13 |
| chr4_177704261_177704412 | 0.56 |
VEGFC |
vascular endothelial growth factor C |
9545 |
0.31 |
| chr11_12221922_12222073 | 0.56 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
38294 |
0.17 |
| chr9_38682670_38682821 | 0.56 |
ENSG00000252725 |
. |
2742 |
0.34 |
| chr15_31676742_31676893 | 0.55 |
KLF13 |
Kruppel-like factor 13 |
18460 |
0.28 |
| chr22_30175136_30175287 | 0.55 |
UQCR10 |
ubiquinol-cytochrome c reductase, complex III subunit X |
11848 |
0.12 |
| chr10_80831884_80832062 | 0.55 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
3181 |
0.3 |
| chr4_20268163_20268314 | 0.55 |
SLIT2 |
slit homolog 2 (Drosophila) |
11695 |
0.31 |
| chr1_44249696_44249884 | 0.55 |
ST3GAL3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
47856 |
0.11 |
| chr1_214741452_214741603 | 0.55 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
16961 |
0.24 |
| chr3_12960579_12960730 | 0.55 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
48514 |
0.12 |
| chr8_62602534_62602708 | 0.55 |
ASPH |
aspartate beta-hydroxylase |
213 |
0.95 |
| chr20_34457569_34457720 | 0.55 |
ENSG00000199676 |
. |
16358 |
0.13 |
| chr8_126496676_126496827 | 0.55 |
ENSG00000266452 |
. |
39944 |
0.18 |
| chr14_56800333_56800607 | 0.55 |
TMEM260 |
transmembrane protein 260 |
154602 |
0.04 |
| chr5_54207045_54207196 | 0.54 |
RP11-45H22.3 |
Uncharacterized protein |
46554 |
0.13 |
| chr6_5125637_5125928 | 0.54 |
ENSG00000265083 |
. |
22774 |
0.19 |
| chr20_50248125_50248545 | 0.54 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
68965 |
0.12 |
| chr5_148346010_148346161 | 0.54 |
RP11-44B19.1 |
|
3818 |
0.26 |
| chr2_74787629_74787875 | 0.54 |
LOXL3 |
lysyl oxidase-like 3 |
4935 |
0.1 |
| chr1_117690445_117690596 | 0.54 |
TRIM45 |
tripartite motif containing 45 |
25311 |
0.15 |
| chr9_97563146_97563297 | 0.54 |
C9orf3 |
chromosome 9 open reading frame 3 |
763 |
0.65 |
| chr19_2543854_2544412 | 0.54 |
ENSG00000252962 |
. |
40994 |
0.11 |
| chr5_148515097_148515248 | 0.53 |
ABLIM3 |
actin binding LIM protein family, member 3 |
5874 |
0.19 |
| chr10_14519559_14519710 | 0.53 |
FRMD4A |
FERM domain containing 4A |
15493 |
0.21 |
| chr17_13447545_13447833 | 0.52 |
ENSG00000221698 |
. |
726 |
0.71 |
| chr8_128403450_128403601 | 0.52 |
POU5F1B |
POU class 5 homeobox 1B |
23010 |
0.23 |
| chr17_62660365_62660516 | 0.52 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
2254 |
0.34 |
| chr18_26734289_26734440 | 0.52 |
ENSG00000212085 |
. |
42128 |
0.18 |
| chr22_36181419_36181847 | 0.52 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
7554 |
0.27 |
| chr20_33166516_33166667 | 0.51 |
MAP1LC3A |
microtubule-associated protein 1 light chain 3 alpha |
20069 |
0.16 |
| chr7_23470661_23470850 | 0.51 |
ENSG00000252590 |
. |
19522 |
0.16 |
| chr20_33895079_33895258 | 0.51 |
FAM83C |
family with sequence similarity 83, member C |
14964 |
0.1 |
| chr22_44453249_44453400 | 0.51 |
PARVB |
parvin, beta |
11617 |
0.25 |
| chr5_67289012_67289163 | 0.51 |
ENSG00000223149 |
. |
25814 |
0.26 |
| chr15_101717639_101717898 | 0.50 |
CHSY1 |
chondroitin sulfate synthase 1 |
74369 |
0.09 |
| chr1_222164593_222164744 | 0.50 |
ENSG00000212094 |
. |
21764 |
0.28 |
| chr20_22775413_22775564 | 0.50 |
ENSG00000265151 |
. |
61549 |
0.14 |
| chr16_67951318_67951477 | 0.50 |
CTRL |
chymotrypsin-like |
14368 |
0.08 |
| chr2_173290792_173290943 | 0.50 |
ITGA6 |
integrin, alpha 6 |
1215 |
0.44 |
| chr11_61463037_61463336 | 0.50 |
DAGLA |
diacylglycerol lipase, alpha |
15281 |
0.16 |
| chr3_69811903_69812159 | 0.50 |
MITF |
microphthalmia-associated transcription factor |
156 |
0.97 |
| chr8_117702787_117702938 | 0.50 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
65161 |
0.12 |
| chr1_19271200_19271351 | 0.49 |
IFFO2 |
intermediate filament family orphan 2 |
10917 |
0.19 |
| chr8_124281675_124281846 | 0.49 |
ZHX1-C8ORF76 |
ZHX1-C8ORF76 readthrough |
2133 |
0.17 |
| chr11_110668037_110668323 | 0.49 |
ARHGAP20 |
Rho GTPase activating protein 20 |
84268 |
0.11 |
| chr6_135174576_135174727 | 0.49 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
75648 |
0.11 |
| chr15_49069969_49070120 | 0.48 |
RP11-485O10.2 |
|
5343 |
0.21 |
| chr1_192499348_192499877 | 0.48 |
RP5-1011O1.2 |
|
36735 |
0.16 |
| chr7_7593210_7593361 | 0.48 |
MIOS |
missing oocyte, meiosis regulator, homolog (Drosophila) |
13218 |
0.22 |
| chr8_127833738_127833889 | 0.48 |
ENSG00000212451 |
. |
150046 |
0.04 |
| chr15_49073804_49073955 | 0.48 |
RP11-485O10.2 |
|
1508 |
0.4 |
| chr7_42717076_42717227 | 0.48 |
C7orf25 |
chromosome 7 open reading frame 25 |
234358 |
0.02 |
| chr1_115723224_115723375 | 0.47 |
TSPAN2 |
tetraspanin 2 |
91178 |
0.08 |
| chr19_43954759_43955038 | 0.47 |
LYPD3 |
LY6/PLAUR domain containing 3 |
14914 |
0.13 |
| chr4_148703798_148703949 | 0.47 |
ENSG00000264274 |
. |
127 |
0.96 |
| chr1_21024911_21025062 | 0.47 |
KIF17 |
kinesin family member 17 |
18998 |
0.13 |
| chr20_11176893_11177136 | 0.47 |
C20orf187 |
chromosome 20 open reading frame 187 |
168203 |
0.04 |
| chr1_40423957_40424317 | 0.47 |
MFSD2A |
major facilitator superfamily domain containing 2A |
3315 |
0.2 |
| chr7_134604064_134604215 | 0.47 |
CALD1 |
caldesmon 1 |
1098 |
0.63 |
| chr7_29149222_29149373 | 0.47 |
CPVL |
carboxypeptidase, vitellogenic-like |
3240 |
0.22 |
| chr15_74373495_74373794 | 0.47 |
GOLGA6A |
golgin A6 family, member A |
1247 |
0.38 |
| chr20_23426178_23426329 | 0.47 |
CSTL1 |
cystatin-like 1 |
5368 |
0.13 |
| chr10_5594332_5594536 | 0.47 |
CALML3-AS1 |
CALML3 antisense RNA 1 |
26225 |
0.13 |
| chr19_612185_612336 | 0.46 |
AC005559.2 |
|
4899 |
0.1 |
| chr13_21607464_21607818 | 0.46 |
ENSG00000201821 |
. |
5165 |
0.17 |
| chr12_54590925_54591203 | 0.46 |
SMUG1 |
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
8286 |
0.12 |
| chr10_133110720_133110871 | 0.46 |
TCERG1L |
transcription elongation regulator 1-like |
811 |
0.78 |
| chr14_37466179_37466330 | 0.46 |
ENSG00000266327 |
. |
44658 |
0.17 |
| chr1_152141043_152141216 | 0.46 |
FLG-AS1 |
FLG antisense RNA 1 |
528 |
0.73 |
| chr1_246020674_246020825 | 0.46 |
RP11-83A16.1 |
|
176104 |
0.03 |
| chr3_141866576_141866846 | 0.46 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
1591 |
0.36 |
| chr6_82543726_82543877 | 0.46 |
ENSG00000206886 |
. |
70060 |
0.11 |
| chr11_85453398_85453549 | 0.45 |
SYTL2 |
synaptotagmin-like 2 |
14402 |
0.19 |
| chr9_42854346_42854549 | 0.45 |
AC129778.2 |
|
1845 |
0.47 |
| chr14_68610729_68611179 | 0.45 |
ENSG00000244677 |
. |
5671 |
0.25 |
| chr1_31575614_31575797 | 0.45 |
PUM1 |
pumilio RNA-binding family member 1 |
36867 |
0.17 |
| chr20_45947327_45947866 | 0.45 |
AL031666.2 |
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
350 |
0.82 |
| chr17_47527935_47528264 | 0.45 |
ENSG00000207127 |
. |
33887 |
0.1 |
| chr2_47139549_47139700 | 0.45 |
MCFD2 |
multiple coagulation factor deficiency 2 |
1589 |
0.3 |
| chr4_114390008_114390159 | 0.45 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
44915 |
0.14 |
| chr13_38069587_38069738 | 0.45 |
POSTN |
periostin, osteoblast specific factor |
103201 |
0.08 |
| chr8_95239241_95239392 | 0.45 |
CDH17 |
cadherin 17, LI cadherin (liver-intestine) |
9785 |
0.24 |
| chr9_69654789_69655001 | 0.44 |
AL445665.1 |
Protein LOC100996643 |
4632 |
0.3 |
| chr19_41727336_41727487 | 0.44 |
CTD-2195B23.3 |
|
545 |
0.64 |
| chr10_61942717_61942868 | 0.44 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
16138 |
0.27 |
| chr7_2146247_2146398 | 0.44 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
1021 |
0.66 |
| chr9_67027339_67027528 | 0.44 |
ENSG00000265285 |
. |
67500 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.1 | 0.5 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.1 | 0.3 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.2 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.5 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.2 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.2 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.1 | 0.2 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.2 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.1 | 0.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.2 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.2 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0044557 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.2 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.0 | 0.1 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.2 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.3 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.0 | GO:0061043 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0031065 | regulation of histone deacetylation(GO:0031063) positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:1904376 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.0 | GO:2000696 | nephron tubule epithelial cell differentiation(GO:0072160) regulation of nephron tubule epithelial cell differentiation(GO:0072182) regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.1 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.0 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0048668 | collateral sprouting(GO:0048668) regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0071636 | positive regulation of transforming growth factor beta production(GO:0071636) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:1902305 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.0 | 0.2 | GO:0035338 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.1 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0034393 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0007435 | salivary gland morphogenesis(GO:0007435) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.0 | GO:0021794 | thalamus development(GO:0021794) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.0 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0031082 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 0.5 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.1 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.5 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.1 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.0 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.0 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.0 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.0 | GO:0047115 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0016531 | metallochaperone activity(GO:0016530) copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | Genes involved in MAPK targets/ Nuclear events mediated by MAP kinases |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |