Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
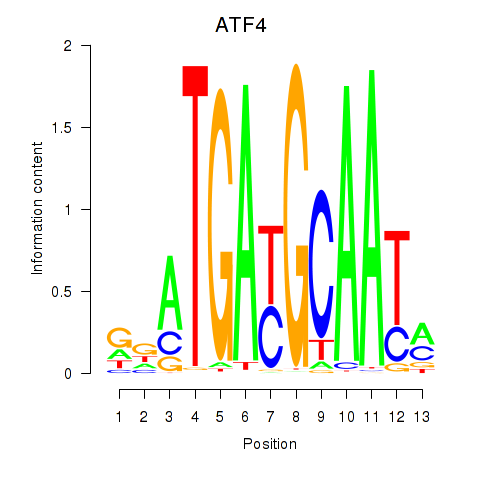
Results for ATF4
Z-value: 1.63
Transcription factors associated with ATF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF4
|
ENSG00000128272.10 | activating transcription factor 4 |
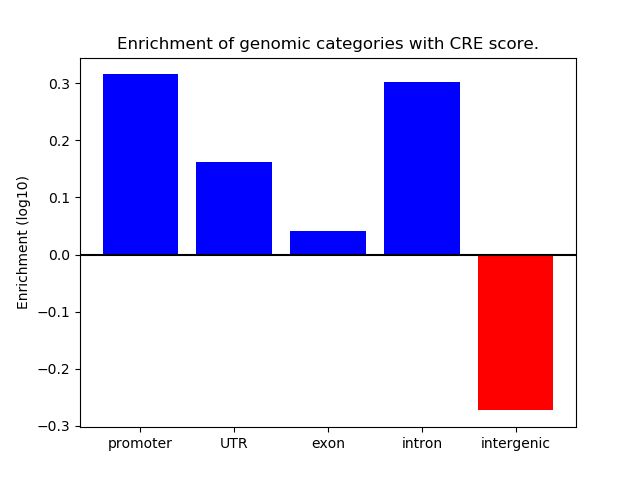
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr22_39920267_39920552 | ATF4 | 3840 | 0.169811 | -0.78 | 1.3e-02 | Click! |
| chr22_39914272_39914423 | ATF4 | 1353 | 0.346581 | 0.73 | 2.5e-02 | Click! |
| chr22_39918738_39919296 | ATF4 | 2448 | 0.215052 | -0.70 | 3.5e-02 | Click! |
| chr22_39914606_39914905 | ATF4 | 945 | 0.478059 | 0.57 | 1.1e-01 | Click! |
| chr22_39918482_39918719 | ATF4 | 2031 | 0.246137 | -0.57 | 1.1e-01 | Click! |
Activity of the ATF4 motif across conditions
Conditions sorted by the z-value of the ATF4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
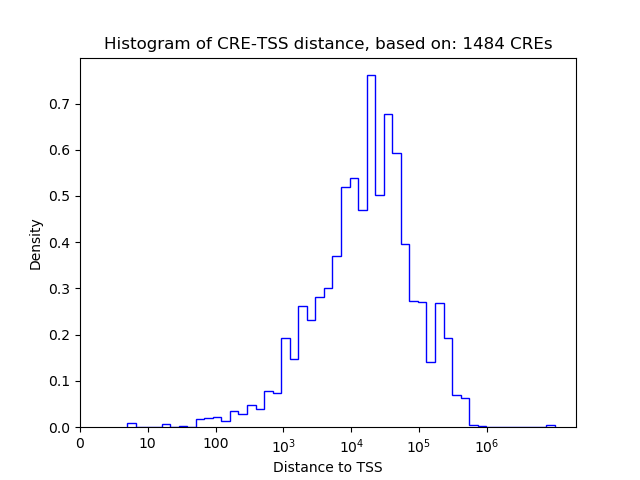
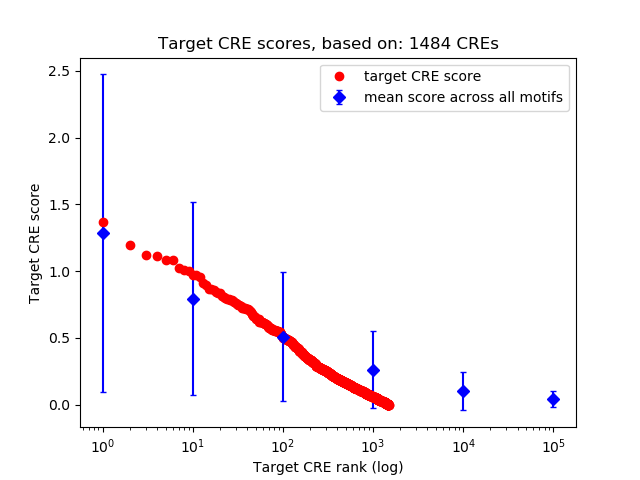
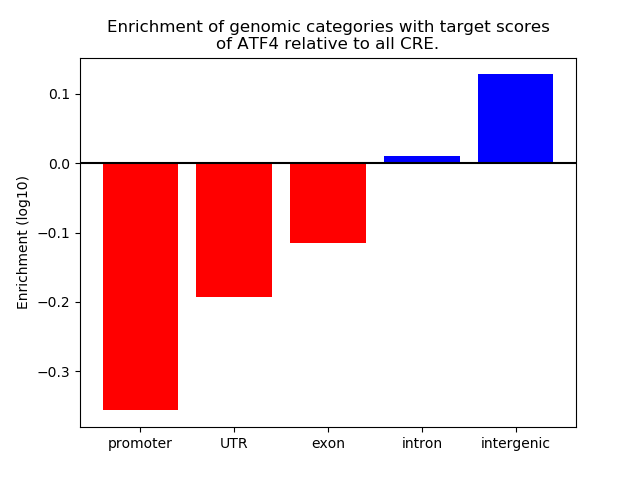
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_30333675_30334003 | 1.37 |
ENSG00000199927 |
. |
12188 |
0.3 |
| chr2_119495850_119496001 | 1.20 |
EN1 |
engrailed homeobox 1 |
109329 |
0.07 |
| chr9_124051991_124052278 | 1.12 |
GSN |
gelsolin |
3264 |
0.18 |
| chr2_55327280_55327923 | 1.12 |
ENSG00000266376 |
. |
8245 |
0.2 |
| chr18_24326732_24327016 | 1.09 |
ENSG00000265369 |
. |
57381 |
0.13 |
| chr20_43124043_43124194 | 1.08 |
SERINC3 |
serine incorporator 3 |
9414 |
0.12 |
| chr10_65462909_65463060 | 1.02 |
REEP3 |
receptor accessory protein 3 |
181861 |
0.03 |
| chr4_86953260_86953440 | 1.01 |
RP13-514E23.1 |
|
18323 |
0.19 |
| chr21_15734345_15734716 | 1.00 |
HSPA13 |
heat shock protein 70kDa family, member 13 |
20928 |
0.24 |
| chr15_71119079_71119471 | 0.97 |
RP11-138H8.6 |
|
22834 |
0.14 |
| chr10_99299975_99300262 | 0.97 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
32080 |
0.09 |
| chr9_93102862_93103013 | 0.96 |
DIRAS2 |
DIRAS family, GTP-binding RAS-like 2 |
302449 |
0.01 |
| chr4_26624592_26624880 | 0.91 |
ENSG00000265627 |
. |
14376 |
0.23 |
| chr16_72909394_72909957 | 0.90 |
ENSG00000251868 |
. |
53784 |
0.12 |
| chr15_36626292_36626443 | 0.87 |
C15orf41 |
chromosome 15 open reading frame 41 |
245445 |
0.02 |
| chr1_8272197_8272744 | 0.87 |
ENSG00000200975 |
. |
5813 |
0.24 |
| chr3_105073937_105074088 | 0.86 |
ALCAM |
activated leukocyte cell adhesion molecule |
11741 |
0.33 |
| chr4_39152584_39152984 | 0.84 |
RP11-360F5.1 |
|
24346 |
0.17 |
| chr4_13920319_13920470 | 0.84 |
ENSG00000252092 |
. |
260143 |
0.02 |
| chr1_204381057_204381208 | 0.83 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
213 |
0.94 |
| chr20_23238843_23239098 | 0.81 |
NXT1 |
NTF2-like export factor 1 |
92403 |
0.05 |
| chr12_52077625_52077776 | 0.81 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
2507 |
0.37 |
| chr4_57887081_57887232 | 0.80 |
ENSG00000251703 |
. |
18298 |
0.15 |
| chr1_221490047_221490325 | 0.79 |
DUSP10 |
dual specificity phosphatase 10 |
420616 |
0.01 |
| chr10_126734388_126734600 | 0.79 |
ENSG00000264572 |
. |
13055 |
0.21 |
| chr8_94910083_94910336 | 0.79 |
ENSG00000264448 |
. |
18138 |
0.18 |
| chr7_98000771_98000922 | 0.78 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
29534 |
0.17 |
| chr11_76288782_76288933 | 0.78 |
C11orf30 |
chromosome 11 open reading frame 30 |
31865 |
0.17 |
| chr6_18216555_18216818 | 0.77 |
KDM1B |
lysine (K)-specific demethylase 1B |
30667 |
0.15 |
| chr20_12997924_12998210 | 0.76 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
8070 |
0.32 |
| chr1_30995259_30995410 | 0.75 |
MATN1-AS1 |
MATN1 antisense RNA 1 |
196017 |
0.03 |
| chr2_118881899_118882337 | 0.75 |
INSIG2 |
insulin induced gene 2 |
36068 |
0.19 |
| chr7_111724278_111724664 | 0.75 |
DOCK4 |
dedicator of cytokinesis 4 |
80285 |
0.1 |
| chr16_8623102_8623253 | 0.74 |
TMEM114 |
transmembrane protein 114 |
951 |
0.68 |
| chr19_2524912_2525063 | 0.73 |
ENSG00000252962 |
. |
21848 |
0.14 |
| chr13_80350766_80350917 | 0.72 |
NDFIP2 |
Nedd4 family interacting protein 2 |
295253 |
0.01 |
| chr9_84106067_84106472 | 0.72 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
122168 |
0.06 |
| chr16_9166899_9167138 | 0.72 |
C16orf72 |
chromosome 16 open reading frame 72 |
18487 |
0.16 |
| chr15_60172096_60172247 | 0.72 |
FOXB1 |
forkhead box B1 |
124250 |
0.05 |
| chr17_76325332_76325654 | 0.71 |
SOCS3 |
suppressor of cytokine signaling 3 |
30662 |
0.12 |
| chr3_149812034_149812185 | 0.71 |
RP11-167H9.4 |
|
2121 |
0.33 |
| chr3_141202423_141202574 | 0.71 |
RASA2 |
RAS p21 protein activator 2 |
3393 |
0.28 |
| chr9_127154555_127154765 | 0.70 |
PSMB7 |
proteasome (prosome, macropain) subunit, beta type, 7 |
23058 |
0.17 |
| chr6_146680396_146680547 | 0.70 |
ENSG00000222971 |
. |
18915 |
0.29 |
| chr8_8457538_8457689 | 0.68 |
ENSG00000263616 |
. |
17448 |
0.22 |
| chr10_74226369_74226629 | 0.68 |
RP11-167P22.3 |
|
35005 |
0.15 |
| chr4_160195981_160196189 | 0.66 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
7196 |
0.24 |
| chr12_44241149_44241300 | 0.66 |
TMEM117 |
transmembrane protein 117 |
2557 |
0.34 |
| chr3_9464516_9464980 | 0.65 |
ENSG00000266001 |
. |
3286 |
0.2 |
| chr6_36635004_36635155 | 0.64 |
ENSG00000251864 |
. |
5536 |
0.15 |
| chr4_170192796_170193656 | 0.64 |
SH3RF1 |
SH3 domain containing ring finger 1 |
970 |
0.68 |
| chr10_79315415_79315652 | 0.64 |
ENSG00000199592 |
. |
31274 |
0.22 |
| chr2_9679702_9679882 | 0.64 |
ENSG00000238462 |
. |
7318 |
0.17 |
| chr10_59784388_59784539 | 0.64 |
ENSG00000238970 |
. |
214058 |
0.02 |
| chr5_73834033_73834384 | 0.62 |
HEXB |
hexosaminidase B (beta polypeptide) |
101640 |
0.07 |
| chr11_63440539_63440690 | 0.62 |
ATL3 |
atlastin GTPase 3 |
1221 |
0.4 |
| chr15_71118369_71118520 | 0.62 |
RP11-138H8.6 |
|
23665 |
0.14 |
| chr2_29287035_29287186 | 0.62 |
C2orf71 |
chromosome 2 open reading frame 71 |
10017 |
0.14 |
| chr1_207489899_207490187 | 0.62 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
4810 |
0.3 |
| chr1_88319838_88319989 | 0.61 |
ENSG00000199318 |
. |
400857 |
0.01 |
| chr1_61629231_61629382 | 0.61 |
RP4-802A10.1 |
|
38901 |
0.19 |
| chr6_130690312_130690483 | 0.61 |
TMEM200A |
transmembrane protein 200A |
3518 |
0.26 |
| chr3_149812224_149812522 | 0.61 |
RP11-167H9.4 |
|
1857 |
0.36 |
| chr11_33715982_33716133 | 0.61 |
RP4-541C22.5 |
|
2190 |
0.27 |
| chr5_96190924_96191075 | 0.60 |
CTD-2260A17.2 |
Uncharacterized protein |
18316 |
0.14 |
| chr7_28102484_28102649 | 0.60 |
JAZF1 |
JAZF zinc finger 1 |
8737 |
0.26 |
| chr2_179279617_179280113 | 0.60 |
AC009948.5 |
|
1115 |
0.47 |
| chr21_34090483_34090634 | 0.59 |
SYNJ1 |
synaptojanin 1 |
8648 |
0.16 |
| chr3_73815335_73815486 | 0.59 |
PDZRN3 |
PDZ domain containing ring finger 3 |
141319 |
0.05 |
| chr2_64017553_64017895 | 0.58 |
UGP2 |
UDP-glucose pyrophosphorylase 2 |
50350 |
0.15 |
| chr15_67799757_67800110 | 0.58 |
C15orf61 |
chromosome 15 open reading frame 61 |
13473 |
0.2 |
| chr16_15698493_15698743 | 0.57 |
CTB-193M12.1 |
|
3713 |
0.19 |
| chr10_65227481_65227632 | 0.57 |
JMJD1C |
jumonji domain containing 1C |
1834 |
0.34 |
| chr6_56634837_56634988 | 0.57 |
DST |
dystonin |
15765 |
0.26 |
| chr4_13921909_13922060 | 0.56 |
ENSG00000252092 |
. |
261733 |
0.02 |
| chr22_41776460_41776636 | 0.56 |
TEF |
thyrotrophic embryonic factor |
1385 |
0.34 |
| chr3_71582773_71583007 | 0.56 |
ENSG00000221264 |
. |
8350 |
0.21 |
| chr17_70808576_70808727 | 0.56 |
ENSG00000252274 |
. |
6460 |
0.22 |
| chr1_234639139_234639392 | 0.56 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
24416 |
0.19 |
| chr10_44338826_44338977 | 0.56 |
ENSG00000238957 |
. |
191087 |
0.03 |
| chr12_89728661_89728921 | 0.56 |
DUSP6 |
dual specificity phosphatase 6 |
16157 |
0.24 |
| chr7_101498442_101498914 | 0.56 |
CTA-339C12.1 |
|
30669 |
0.18 |
| chr2_174016909_174017060 | 0.55 |
MLK7-AS1 |
MLK7 antisense RNA 1 |
55356 |
0.13 |
| chr5_73092459_73092753 | 0.55 |
CTC-575I10.1 |
|
10290 |
0.2 |
| chr5_81644909_81645060 | 0.55 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
43818 |
0.19 |
| chr10_22284671_22284822 | 0.55 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
7582 |
0.28 |
| chr2_47140554_47140705 | 0.55 |
MCFD2 |
multiple coagulation factor deficiency 2 |
584 |
0.67 |
| chr10_91106448_91106732 | 0.55 |
LIPA |
lipase A, lysosomal acid, cholesterol esterase |
4129 |
0.16 |
| chr2_228681785_228681989 | 0.55 |
CCL20 |
chemokine (C-C motif) ligand 20 |
3317 |
0.27 |
| chr6_5206187_5206422 | 0.55 |
ENSG00000264541 |
. |
18726 |
0.19 |
| chr6_13762651_13763628 | 0.54 |
MCUR1 |
mitochondrial calcium uniporter regulator 1 |
38438 |
0.15 |
| chr1_152001823_152002025 | 0.54 |
S100A11 |
S100 calcium binding protein A11 |
7587 |
0.15 |
| chr1_180092706_180092857 | 0.54 |
CEP350 |
centrosomal protein 350kDa |
29932 |
0.18 |
| chr13_43407082_43407233 | 0.54 |
FAM216B |
family with sequence similarity 216, member B |
51406 |
0.18 |
| chr2_13058685_13058836 | 0.53 |
ENSG00000264370 |
. |
181267 |
0.03 |
| chr20_61303299_61303450 | 0.53 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
4209 |
0.14 |
| chr13_96408525_96408676 | 0.52 |
ENSG00000251901 |
. |
41568 |
0.18 |
| chr3_138582119_138582540 | 0.51 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
28549 |
0.2 |
| chr10_75652450_75652601 | 0.51 |
PLAU |
plasminogen activator, urokinase |
16410 |
0.11 |
| chr13_110033704_110033855 | 0.51 |
MYO16-AS1 |
MYO16 antisense RNA 1 |
179948 |
0.03 |
| chr6_23175706_23175857 | 0.50 |
ENSG00000207394 |
. |
50466 |
0.2 |
| chr7_129338898_129339049 | 0.50 |
NRF1 |
nuclear respiratory factor 1 |
41782 |
0.13 |
| chr21_36168084_36168235 | 0.50 |
AP000330.8 |
|
50025 |
0.16 |
| chr3_172345080_172345231 | 0.50 |
AC007919.2 |
HCG1787166; PRO1163; Uncharacterized protein |
16328 |
0.19 |
| chr5_16736411_16736625 | 0.50 |
MYO10 |
myosin X |
1978 |
0.43 |
| chr4_54373291_54373442 | 0.49 |
LNX1-AS1 |
LNX1 antisense RNA 1 |
5308 |
0.25 |
| chr6_149333665_149333816 | 0.49 |
RP11-162J8.3 |
|
19969 |
0.23 |
| chr1_33453155_33453306 | 0.49 |
RP1-117O3.2 |
|
554 |
0.7 |
| chr2_121507260_121507411 | 0.49 |
GLI2 |
GLI family zinc finger 2 |
13512 |
0.29 |
| chr17_36571605_36571871 | 0.49 |
ENSG00000266103 |
. |
5696 |
0.17 |
| chr10_33604953_33605104 | 0.49 |
NRP1 |
neuropilin 1 |
18282 |
0.22 |
| chr11_58394708_58394859 | 0.49 |
CNTF |
ciliary neurotrophic factor |
4637 |
0.18 |
| chr2_205441256_205441407 | 0.48 |
PARD3B |
par-3 family cell polarity regulator beta |
30608 |
0.27 |
| chr5_138517708_138518085 | 0.48 |
SIL1 |
SIL1 nucleotide exchange factor |
14314 |
0.18 |
| chr7_44298814_44299210 | 0.48 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
17107 |
0.18 |
| chr11_102097026_102097177 | 0.48 |
RP11-864G5.3 |
|
4552 |
0.23 |
| chr12_19395564_19395847 | 0.48 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
5847 |
0.26 |
| chr4_102023208_102023359 | 0.48 |
ENSG00000221265 |
. |
228288 |
0.02 |
| chr8_123651385_123651590 | 0.47 |
ENSG00000238901 |
. |
32043 |
0.23 |
| chr8_38483788_38483951 | 0.47 |
C8orf86 |
chromosome 8 open reading frame 86 |
97689 |
0.06 |
| chr1_64211693_64212078 | 0.47 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
27808 |
0.21 |
| chr11_20473321_20473556 | 0.47 |
PRMT3 |
protein arginine methyltransferase 3 |
64176 |
0.13 |
| chr5_136899404_136899555 | 0.47 |
ENSG00000221612 |
. |
30336 |
0.19 |
| chr18_434239_434644 | 0.47 |
RP11-720L2.2 |
|
10025 |
0.23 |
| chr5_118733387_118734036 | 0.47 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
41981 |
0.13 |
| chr6_14728915_14729066 | 0.46 |
ENSG00000206960 |
. |
82224 |
0.11 |
| chr1_222628880_222629091 | 0.46 |
ENSG00000222399 |
. |
48092 |
0.16 |
| chr4_146764367_146764859 | 0.46 |
RP11-181K12.2 |
|
10343 |
0.26 |
| chr11_46016222_46016373 | 0.46 |
PHF21A |
PHD finger protein 21A |
48877 |
0.11 |
| chr3_171685115_171685266 | 0.45 |
FNDC3B |
fibronectin type III domain containing 3B |
72228 |
0.1 |
| chr1_184758212_184758610 | 0.45 |
FAM129A |
family with sequence similarity 129, member A |
16708 |
0.19 |
| chr9_37995152_37995751 | 0.45 |
ENSG00000251745 |
. |
58686 |
0.12 |
| chr5_137854815_137854966 | 0.45 |
ETF1 |
eukaryotic translation termination factor 1 |
22510 |
0.13 |
| chr3_58045428_58045645 | 0.45 |
FLNB |
filamin B, beta |
18520 |
0.24 |
| chr12_28692903_28693202 | 0.44 |
CCDC91 |
coiled-coil domain containing 91 |
8944 |
0.27 |
| chr14_53320261_53320477 | 0.44 |
FERMT2 |
fermitin family member 2 |
10870 |
0.21 |
| chr22_38568821_38569004 | 0.43 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
7499 |
0.14 |
| chr19_31807413_31807564 | 0.43 |
AC007796.1 |
|
32299 |
0.2 |
| chr6_138225502_138226170 | 0.43 |
RP11-356I2.4 |
|
36466 |
0.18 |
| chr18_42900853_42901004 | 0.43 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
107968 |
0.06 |
| chr13_110572258_110572409 | 0.43 |
ENSG00000201161 |
. |
4890 |
0.33 |
| chr7_81589929_81590080 | 0.43 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
10401 |
0.24 |
| chr18_46074818_46074969 | 0.43 |
CTIF |
CBP80/20-dependent translation initiation factor |
8427 |
0.26 |
| chr2_208176087_208176546 | 0.43 |
ENSG00000221628 |
. |
42168 |
0.17 |
| chr14_51471507_51471658 | 0.42 |
PYGL |
phosphorylase, glycogen, liver |
60128 |
0.1 |
| chr10_79021361_79021706 | 0.42 |
RP11-328K22.1 |
|
52166 |
0.15 |
| chr20_19051967_19052205 | 0.42 |
ENSG00000264669 |
. |
54343 |
0.15 |
| chr11_9818939_9819090 | 0.42 |
SBF2 |
SET binding factor 2 |
8263 |
0.17 |
| chr14_85983583_85983734 | 0.41 |
RP11-497E19.2 |
Uncharacterized protein |
11285 |
0.25 |
| chr1_66277924_66278361 | 0.41 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
19278 |
0.24 |
| chr2_162969937_162970139 | 0.40 |
GCG |
glucagon |
38875 |
0.13 |
| chr11_9236582_9237086 | 0.40 |
RP11-5L12.1 |
|
30222 |
0.16 |
| chr6_41065364_41065515 | 0.40 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
58 |
0.96 |
| chr3_99721724_99721891 | 0.40 |
ENSG00000264897 |
. |
38565 |
0.19 |
| chr2_145203456_145203749 | 0.40 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
15465 |
0.27 |
| chr1_63142438_63142826 | 0.40 |
DOCK7 |
dedicator of cytokinesis 7 |
11321 |
0.2 |
| chr12_67927970_67928121 | 0.39 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
114073 |
0.07 |
| chr6_7058768_7059017 | 0.39 |
ENSG00000251762 |
. |
18113 |
0.22 |
| chr4_151070613_151070764 | 0.39 |
ENSG00000238721 |
. |
60170 |
0.14 |
| chr11_32421982_32422133 | 0.39 |
WT1 |
Wilms tumor 1 |
104 |
0.98 |
| chr8_28565507_28565705 | 0.39 |
EXTL3 |
exostosin-like glycosyltransferase 3 |
2269 |
0.3 |
| chr4_48915724_48915875 | 0.39 |
OCIAD2 |
OCIA domain containing 2 |
6954 |
0.2 |
| chr11_57045647_57045894 | 0.39 |
APLNR |
apelin receptor |
41095 |
0.09 |
| chr8_124179944_124180219 | 0.38 |
FAM83A |
family with sequence similarity 83, member A |
11206 |
0.12 |
| chr3_50704589_50704740 | 0.38 |
ENSG00000272543 |
. |
7847 |
0.16 |
| chr5_14305370_14305521 | 0.38 |
TRIO |
trio Rho guanine nucleotide exchange factor |
14359 |
0.31 |
| chr2_190118686_190118837 | 0.38 |
ENSG00000266817 |
. |
57726 |
0.13 |
| chr8_116461452_116461961 | 0.38 |
TRPS1 |
trichorhinophalangeal syndrome I |
42742 |
0.19 |
| chr18_53400073_53400224 | 0.38 |
TCF4 |
transcription factor 4 |
68130 |
0.14 |
| chr4_8527517_8527668 | 0.37 |
GPR78 |
G protein-coupled receptor 78 |
54701 |
0.11 |
| chr11_74031798_74031949 | 0.37 |
P4HA3 |
prolyl 4-hydroxylase, alpha polypeptide III |
9171 |
0.18 |
| chr8_26469143_26469294 | 0.37 |
DPYSL2 |
dihydropyrimidinase-like 2 |
33297 |
0.21 |
| chr1_198227986_198228137 | 0.37 |
NEK7 |
NIMA-related kinase 7 |
38132 |
0.23 |
| chr1_6230966_6231267 | 0.37 |
CHD5 |
chromodomain helicase DNA binding protein 5 |
9067 |
0.14 |
| chr22_43263821_43263972 | 0.37 |
ARFGAP3 |
ADP-ribosylation factor GTPase activating protein 3 |
9784 |
0.23 |
| chr15_42578664_42578815 | 0.36 |
GANC |
glucosidase, alpha; neutral C |
12299 |
0.16 |
| chr11_129093775_129094069 | 0.36 |
ENSG00000199260 |
. |
21061 |
0.22 |
| chr9_128223375_128223526 | 0.36 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
23330 |
0.21 |
| chr2_46066755_46066906 | 0.36 |
PRKCE |
protein kinase C, epsilon |
161211 |
0.04 |
| chr9_128005655_128005806 | 0.36 |
RP11-65N13.8 |
|
1788 |
0.26 |
| chr1_186782472_186782623 | 0.36 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
15538 |
0.29 |
| chr2_109565955_109566106 | 0.36 |
EDAR |
ectodysplasin A receptor |
39695 |
0.19 |
| chr3_42068290_42068573 | 0.35 |
TRAK1 |
trafficking protein, kinesin binding 1 |
64131 |
0.11 |
| chr11_72680748_72681422 | 0.35 |
FCHSD2 |
FCH and double SH3 domains 2 |
18972 |
0.22 |
| chr1_61586786_61586937 | 0.35 |
RP4-802A10.1 |
|
3544 |
0.28 |
| chr16_48400227_48400867 | 0.35 |
SIAH1 |
siah E3 ubiquitin protein ligase 1 |
735 |
0.66 |
| chr4_108921158_108921309 | 0.35 |
HADH |
hydroxyacyl-CoA dehydrogenase |
4558 |
0.22 |
| chr11_34393224_34393428 | 0.35 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
13771 |
0.25 |
| chr8_129189248_129189490 | 0.35 |
ENSG00000221261 |
. |
27007 |
0.21 |
| chr8_93552671_93552822 | 0.35 |
ENSG00000221172 |
. |
94817 |
0.09 |
| chr15_40200299_40200575 | 0.35 |
GPR176 |
G protein-coupled receptor 176 |
11989 |
0.15 |
| chr6_89744532_89744683 | 0.34 |
ENSG00000223001 |
. |
28802 |
0.13 |
| chr3_131448487_131448638 | 0.34 |
CPNE4 |
copine IV |
195863 |
0.03 |
| chr10_75346559_75346987 | 0.34 |
USP54 |
ubiquitin specific peptidase 54 |
4323 |
0.16 |
| chr14_63940302_63940453 | 0.34 |
ENSG00000252463 |
. |
4115 |
0.18 |
| chr4_178362396_178362599 | 0.34 |
AGA |
aspartylglucosaminidase |
1160 |
0.46 |
| chr9_123495391_123495542 | 0.34 |
MEGF9 |
multiple EGF-like-domains 9 |
18718 |
0.21 |
| chr14_50469321_50469915 | 0.34 |
C14orf182 |
chromosome 14 open reading frame 182 |
4620 |
0.21 |
| chr8_37639065_37639216 | 0.34 |
GPR124 |
G protein-coupled receptor 124 |
2569 |
0.21 |
| chr8_141607678_141608368 | 0.34 |
AGO2 |
argonaute RISC catalytic component 2 |
7964 |
0.25 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.8 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.2 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.1 | 0.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.2 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0072191 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0032347 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0071501 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0048343 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.3 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0014848 | urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.4 | GO:0090659 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.1 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.3 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0046471 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0055057 | neuronal stem cell division(GO:0036445) neuroblast division(GO:0055057) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0051957 | positive regulation of glutamate secretion(GO:0014049) positive regulation of amino acid transport(GO:0051957) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:0043558 | regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 | 0.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.0 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0031394 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.0 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.0 | GO:0032692 | negative regulation of interleukin-1 production(GO:0032692) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.2 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.2 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.0 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0002060 | nucleobase binding(GO:0002054) purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |