Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
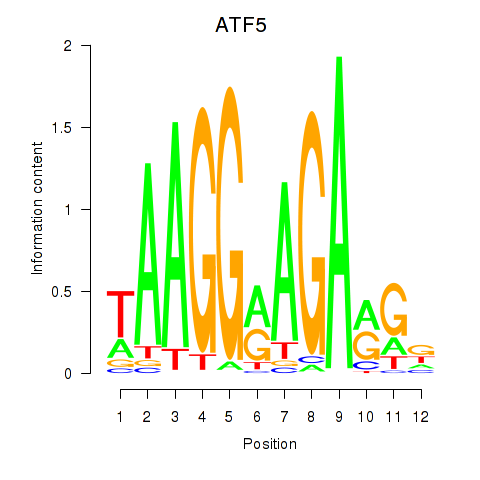
Results for ATF5
Z-value: 1.61
Transcription factors associated with ATF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF5
|
ENSG00000169136.4 | activating transcription factor 5 |
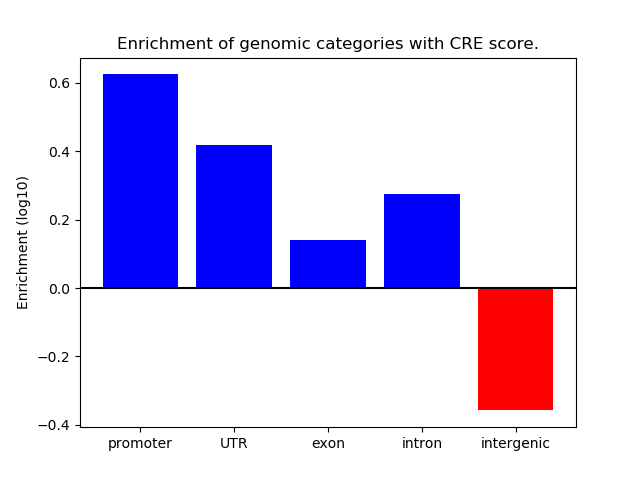
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_50431904_50432210 | ATF5 | 98 | 0.805868 | 0.34 | 3.7e-01 | Click! |
| chr19_50427511_50427662 | ATF5 | 4373 | 0.077943 | -0.29 | 4.5e-01 | Click! |
| chr19_50426496_50426647 | ATF5 | 5388 | 0.073280 | -0.24 | 5.3e-01 | Click! |
| chr19_50427838_50427989 | ATF5 | 4046 | 0.080094 | -0.20 | 6.1e-01 | Click! |
| chr19_50433408_50433958 | ATF5 | 207 | 0.703805 | -0.13 | 7.4e-01 | Click! |
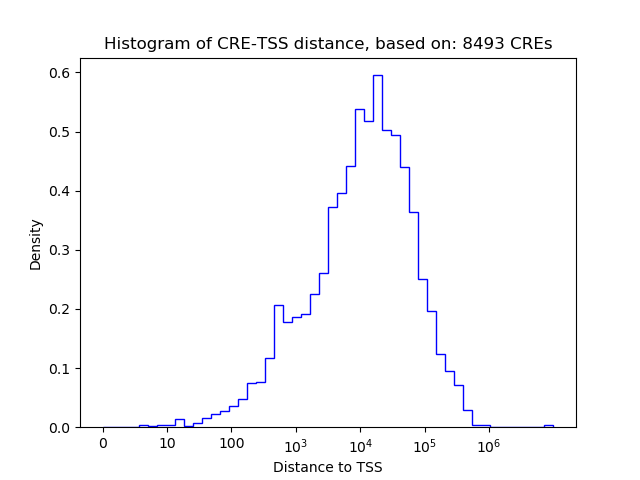
Activity of the ATF5 motif across conditions
Conditions sorted by the z-value of the ATF5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
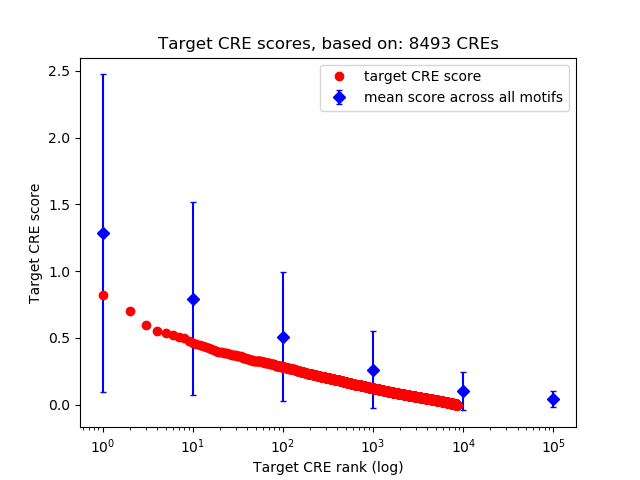
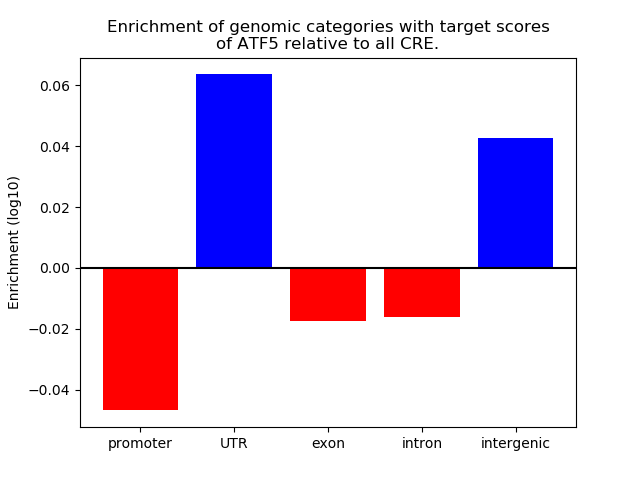
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_194456835_194457137 | 0.82 |
FAM43A |
family with sequence similarity 43, member A |
50364 |
0.11 |
| chr22_37696225_37696376 | 0.70 |
CYTH4 |
cytohesin 4 |
9709 |
0.17 |
| chr12_62691099_62691281 | 0.59 |
USP15 |
ubiquitin specific peptidase 15 |
8186 |
0.22 |
| chr17_14215558_14215761 | 0.55 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
11259 |
0.27 |
| chr5_36724502_36724744 | 0.53 |
CTD-2353F22.1 |
|
652 |
0.82 |
| chr11_9573412_9573568 | 0.52 |
WEE1 |
WEE1 G2 checkpoint kinase |
21738 |
0.13 |
| chr12_106177631_106177796 | 0.51 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
300098 |
0.01 |
| chr17_32706061_32706253 | 0.50 |
CCL1 |
chemokine (C-C motif) ligand 1 |
15907 |
0.17 |
| chr20_43996735_43996900 | 0.48 |
SYS1 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
4723 |
0.13 |
| chr3_177142028_177142201 | 0.46 |
ENSG00000252028 |
. |
79226 |
0.12 |
| chr16_8725990_8726202 | 0.46 |
METTL22 |
methyltransferase like 22 |
8899 |
0.2 |
| chr19_47226837_47226988 | 0.45 |
STRN4 |
striatin, calmodulin binding protein 4 |
444 |
0.67 |
| chr10_98416850_98417114 | 0.44 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
12386 |
0.2 |
| chr20_62492291_62492444 | 0.43 |
ABHD16B |
abhydrolase domain containing 16B |
199 |
0.8 |
| chr8_131217304_131217455 | 0.43 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
24271 |
0.21 |
| chr1_66717796_66717947 | 0.41 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
5459 |
0.33 |
| chr13_41576792_41576957 | 0.41 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
16576 |
0.19 |
| chr18_21589322_21589666 | 0.40 |
TTC39C |
tetratricopeptide repeat domain 39C |
4890 |
0.2 |
| chr1_90222995_90223224 | 0.40 |
ENSG00000239176 |
. |
10768 |
0.21 |
| chr1_36359399_36359646 | 0.40 |
AGO1 |
argonaute RISC catalytic component 1 |
10712 |
0.2 |
| chr9_7011676_7011831 | 0.39 |
KDM4C |
lysine (K)-specific demethylase 4C |
2035 |
0.48 |
| chr9_127455863_127456089 | 0.39 |
ENSG00000207737 |
. |
13 |
0.96 |
| chr13_30950302_30950776 | 0.39 |
KATNAL1 |
katanin p60 subunit A-like 1 |
68918 |
0.11 |
| chr4_82413655_82413934 | 0.39 |
RASGEF1B |
RasGEF domain family, member 1B |
20725 |
0.28 |
| chr10_115082326_115082477 | 0.38 |
ENSG00000238380 |
. |
30783 |
0.24 |
| chrX_65206607_65206882 | 0.38 |
RP6-159A1.3 |
|
12849 |
0.22 |
| chr17_66252375_66252526 | 0.37 |
ARSG |
arylsulfatase G |
2873 |
0.21 |
| chr9_92098673_92099046 | 0.37 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
4054 |
0.27 |
| chr2_145279915_145280210 | 0.37 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
1441 |
0.42 |
| chr9_139947067_139947247 | 0.37 |
RP11-229P13.15 |
|
69 |
0.89 |
| chr1_162287658_162287874 | 0.37 |
RP11-565P22.2 |
|
182 |
0.93 |
| chr8_56837997_56838148 | 0.37 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
14721 |
0.14 |
| chr12_6753605_6754226 | 0.36 |
ACRBP |
acrosin binding protein |
2644 |
0.12 |
| chr17_55063294_55063526 | 0.36 |
RP5-1107A17.4 |
|
3300 |
0.17 |
| chr1_209931882_209932204 | 0.36 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
200 |
0.92 |
| chr5_150599949_150600100 | 0.35 |
CCDC69 |
coiled-coil domain containing 69 |
3682 |
0.22 |
| chr3_119216680_119216943 | 0.35 |
TIMMDC1 |
translocase of inner mitochondrial membrane domain containing 1 |
568 |
0.72 |
| chr16_28505934_28506370 | 0.35 |
APOBR |
apolipoprotein B receptor |
159 |
0.85 |
| chr22_37333070_37333221 | 0.35 |
ENSG00000239056 |
. |
11377 |
0.12 |
| chr3_135219068_135219244 | 0.34 |
ENSG00000253004 |
. |
299466 |
0.01 |
| chr1_113589732_113590143 | 0.34 |
LRIG2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
25894 |
0.22 |
| chr15_70628976_70629127 | 0.34 |
ENSG00000200216 |
. |
143476 |
0.05 |
| chr9_140541223_140541390 | 0.34 |
EHMT1 |
euchromatic histone-lysine N-methyltransferase 1 |
27852 |
0.12 |
| chr3_101504132_101504283 | 0.33 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
37 |
0.97 |
| chr20_4793040_4793193 | 0.33 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
2653 |
0.29 |
| chrX_65222769_65222920 | 0.33 |
RP6-159A1.3 |
|
3251 |
0.27 |
| chr2_74175450_74175659 | 0.33 |
ENSG00000201876 |
. |
19779 |
0.14 |
| chr14_35183268_35183419 | 0.33 |
CFL2 |
cofilin 2 (muscle) |
99 |
0.97 |
| chr4_164529822_164529973 | 0.33 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
4789 |
0.31 |
| chr3_171779828_171780095 | 0.33 |
FNDC3B |
fibronectin type III domain containing 3B |
15258 |
0.27 |
| chr1_209930798_209930949 | 0.33 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
970 |
0.47 |
| chr1_17449628_17449779 | 0.33 |
PADI2 |
peptidyl arginine deiminase, type II |
3755 |
0.22 |
| chrX_17700747_17700898 | 0.33 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
47409 |
0.15 |
| chr12_4548368_4548657 | 0.33 |
FGF6 |
fibroblast growth factor 6 |
4873 |
0.24 |
| chr19_6958874_6959025 | 0.33 |
RP11-1137G4.3 |
|
5727 |
0.16 |
| chr12_63308633_63308784 | 0.33 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
20109 |
0.2 |
| chr7_81404438_81404589 | 0.32 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
4759 |
0.35 |
| chr3_14693666_14694016 | 0.32 |
CCDC174 |
coiled-coil domain containing 174 |
570 |
0.73 |
| chr19_46005187_46005379 | 0.32 |
PPM1N |
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
2064 |
0.19 |
| chr8_8678347_8678544 | 0.32 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
72710 |
0.1 |
| chr8_65776788_65776939 | 0.32 |
CYP7B1 |
cytochrome P450, family 7, subfamily B, polypeptide 1 |
65545 |
0.14 |
| chr9_115995100_115995251 | 0.32 |
SLC31A1 |
solute carrier family 31 (copper transporter), member 1 |
11333 |
0.15 |
| chr19_19271102_19271312 | 0.32 |
MEF2B |
myocyte enhancer factor 2B |
9847 |
0.11 |
| chr1_161180255_161180406 | 0.32 |
FCER1G |
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
4739 |
0.07 |
| chr7_41379726_41379877 | 0.31 |
INHBA-AS1 |
INHBA antisense RNA 1 |
353713 |
0.01 |
| chr1_169597496_169597737 | 0.31 |
SELP |
selectin P (granule membrane protein 140kDa, antigen CD62) |
1698 |
0.41 |
| chr5_177959921_177960072 | 0.31 |
COL23A1 |
collagen, type XXIII, alpha 1 |
29243 |
0.19 |
| chr15_99499703_99499854 | 0.31 |
IGF1R |
insulin-like growth factor 1 receptor |
31949 |
0.16 |
| chr13_43744708_43744859 | 0.31 |
DNAJC15 |
DnaJ (Hsp40) homolog, subfamily C, member 15 |
147444 |
0.05 |
| chr19_52269762_52269913 | 0.31 |
FPR2 |
formyl peptide receptor 2 |
1794 |
0.24 |
| chr4_185819150_185819410 | 0.31 |
ENSG00000263458 |
. |
40314 |
0.14 |
| chr22_17597863_17598182 | 0.31 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
4121 |
0.17 |
| chr1_194039700_194039851 | 0.31 |
ENSG00000252241 |
. |
338701 |
0.01 |
| chr6_135602284_135602577 | 0.31 |
RP3-388E23.2 |
|
20276 |
0.17 |
| chr6_138289503_138289940 | 0.31 |
RP11-356I2.4 |
|
100351 |
0.07 |
| chr2_7570361_7571021 | 0.31 |
ENSG00000221255 |
. |
146281 |
0.05 |
| chr10_106010241_106010392 | 0.30 |
GSTO1 |
glutathione S-transferase omega 1 |
3636 |
0.16 |
| chr12_123284211_123284445 | 0.30 |
CCDC62 |
coiled-coil domain containing 62 |
18541 |
0.13 |
| chr21_43710315_43710466 | 0.30 |
ENSG00000216197 |
. |
10065 |
0.15 |
| chr1_28320130_28320423 | 0.30 |
RP11-460I13.2 |
|
33572 |
0.1 |
| chr10_64519735_64519887 | 0.30 |
ADO |
2-aminoethanethiol (cysteamine) dioxygenase |
44705 |
0.14 |
| chr2_218884404_218884588 | 0.30 |
TNS1 |
tensin 1 |
16778 |
0.16 |
| chrX_147416060_147416261 | 0.30 |
AC002368.4 |
|
165975 |
0.04 |
| chr1_9303502_9303653 | 0.29 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
3674 |
0.24 |
| chr2_232334294_232334692 | 0.29 |
NCL |
nucleolin |
5188 |
0.11 |
| chr10_82355785_82355994 | 0.29 |
SH2D4B |
SH2 domain containing 4B |
55314 |
0.14 |
| chr10_7199881_7200167 | 0.29 |
SFMBT2 |
Scm-like with four mbt domains 2 |
250683 |
0.02 |
| chr1_17394434_17394729 | 0.29 |
SDHB |
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
13916 |
0.17 |
| chr10_134148263_134148414 | 0.29 |
LRRC27 |
leucine rich repeat containing 27 |
274 |
0.91 |
| chr5_156706986_156707255 | 0.29 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
5252 |
0.16 |
| chr3_72679493_72679778 | 0.29 |
ENSG00000199469 |
. |
53793 |
0.13 |
| chr11_127345243_127345408 | 0.29 |
ENSG00000223315 |
. |
67579 |
0.13 |
| chr6_20068674_20068825 | 0.29 |
RP11-239H6.2 |
|
143569 |
0.04 |
| chr7_128709757_128710019 | 0.29 |
ENSG00000238733 |
. |
8303 |
0.17 |
| chr1_169665494_169666108 | 0.29 |
SELL |
selectin L |
15038 |
0.19 |
| chr1_249101157_249101330 | 0.29 |
SH3BP5L |
SH3-binding domain protein 5-like |
10029 |
0.16 |
| chr19_52035107_52035430 | 0.29 |
SIGLEC6 |
sialic acid binding Ig-like lectin 6 |
158 |
0.92 |
| chr6_90793986_90794257 | 0.28 |
ENSG00000222078 |
. |
82896 |
0.09 |
| chrX_70917357_70917654 | 0.28 |
ENSG00000221684 |
. |
61800 |
0.11 |
| chrX_52950600_52950792 | 0.28 |
FAM156B |
family with sequence similarity 156, member B |
22287 |
0.17 |
| chr7_141360008_141360198 | 0.28 |
RP5-894A10.2 |
|
3493 |
0.17 |
| chr16_23870015_23870166 | 0.28 |
PRKCB |
protein kinase C, beta |
21546 |
0.22 |
| chr10_120488698_120489276 | 0.28 |
CACUL1 |
CDK2-associated, cullin domain 1 |
25771 |
0.2 |
| chr3_157161787_157162122 | 0.28 |
PTX3 |
pentraxin 3, long |
7376 |
0.26 |
| chr7_5819044_5819195 | 0.28 |
RNF216 |
ring finger protein 216 |
2132 |
0.31 |
| chr3_17024083_17024265 | 0.28 |
PLCL2 |
phospholipase C-like 2 |
27812 |
0.2 |
| chr9_137826567_137827103 | 0.28 |
FCN1 |
ficolin (collagen/fibrinogen domain containing) 1 |
17026 |
0.14 |
| chr1_111433483_111434153 | 0.28 |
CD53 |
CD53 molecule |
18042 |
0.16 |
| chr2_29223602_29224161 | 0.28 |
FAM179A |
family with sequence similarity 179, member A |
17246 |
0.14 |
| chr21_37884637_37884788 | 0.28 |
AP000695.1 |
|
26547 |
0.16 |
| chr1_160681902_160682094 | 0.28 |
CD48 |
CD48 molecule |
357 |
0.85 |
| chr5_95674050_95674201 | 0.27 |
PCSK1 |
proprotein convertase subtilisin/kexin type 1 |
93739 |
0.08 |
| chr20_31096438_31096678 | 0.27 |
C20orf112 |
chromosome 20 open reading frame 112 |
2652 |
0.27 |
| chr1_206954843_206955128 | 0.27 |
IL10 |
interleukin 10 |
9146 |
0.16 |
| chr2_231526275_231526495 | 0.27 |
CAB39 |
calcium binding protein 39 |
51175 |
0.12 |
| chr2_8678901_8679157 | 0.27 |
AC011747.7 |
|
136867 |
0.05 |
| chrX_70279954_70280194 | 0.27 |
SNX12 |
sorting nexin 12 |
8179 |
0.13 |
| chr17_76907206_76907515 | 0.27 |
CTD-2373H9.5 |
|
8015 |
0.14 |
| chr1_146675568_146675782 | 0.27 |
FMO5 |
flavin containing monooxygenase 5 |
21226 |
0.14 |
| chr16_66264539_66264690 | 0.27 |
ENSG00000201999 |
. |
71098 |
0.11 |
| chr20_47430189_47430589 | 0.27 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
14031 |
0.26 |
| chr17_25857470_25857637 | 0.27 |
KSR1 |
kinase suppressor of ras 1 |
52394 |
0.12 |
| chr6_106896400_106896618 | 0.27 |
ENSG00000202386 |
. |
747 |
0.63 |
| chr20_60487292_60487443 | 0.27 |
ENSG00000221417 |
. |
41351 |
0.17 |
| chr8_21341059_21341347 | 0.27 |
ENSG00000266713 |
. |
10336 |
0.28 |
| chr7_18184166_18184423 | 0.27 |
ENSG00000221393 |
. |
17362 |
0.24 |
| chr1_198633904_198634162 | 0.27 |
RP11-553K8.5 |
|
2157 |
0.39 |
| chr5_39203706_39203857 | 0.27 |
FYB |
FYN binding protein |
652 |
0.81 |
| chr15_74285416_74285684 | 0.27 |
STOML1 |
stomatin (EPB72)-like 1 |
861 |
0.47 |
| chr5_114960864_114961097 | 0.27 |
TMED7-TICAM2 |
TMED7-TICAM2 readthrough |
731 |
0.35 |
| chr1_108430883_108431034 | 0.27 |
VAV3-AS1 |
VAV3 antisense RNA 1 |
76107 |
0.1 |
| chr3_138501187_138501338 | 0.26 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
11865 |
0.27 |
| chr12_51155615_51155794 | 0.26 |
ATF1 |
activating transcription factor 1 |
1789 |
0.35 |
| chr16_88433_88739 | 0.26 |
SNRNP25 |
small nuclear ribonucleoprotein 25kDa (U11/U12) |
14424 |
0.09 |
| chr9_90227526_90227781 | 0.26 |
DAPK1-IT1 |
DAPK1 intronic transcript 1 (non-protein coding) |
59284 |
0.13 |
| chr8_96560843_96561107 | 0.26 |
ENSG00000202112 |
. |
50367 |
0.18 |
| chrX_65220131_65220324 | 0.26 |
RP6-159A1.3 |
|
634 |
0.76 |
| chr21_36401379_36401530 | 0.26 |
RUNX1 |
runt-related transcription factor 1 |
20008 |
0.29 |
| chr6_109162800_109163213 | 0.26 |
ARMC2 |
armadillo repeat containing 2 |
6613 |
0.28 |
| chr7_79736267_79736525 | 0.26 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
26875 |
0.25 |
| chr10_6981008_6981159 | 0.26 |
PRKCQ |
protein kinase C, theta |
358820 |
0.01 |
| chr7_20510929_20511080 | 0.26 |
ENSG00000200753 |
. |
94050 |
0.08 |
| chr7_37381820_37382186 | 0.26 |
ELMO1 |
engulfment and cell motility 1 |
364 |
0.89 |
| chr7_30200789_30200940 | 0.26 |
AC007036.5 |
|
3717 |
0.21 |
| chr2_90067763_90067914 | 0.26 |
IGKV6D-21 |
immunoglobulin kappa variable 6D-21 (non-functional) |
7461 |
0.07 |
| chr7_105087983_105088187 | 0.25 |
SRPK2 |
SRSF protein kinase 2 |
58247 |
0.11 |
| chr10_6604152_6604623 | 0.25 |
PRKCQ |
protein kinase C, theta |
17814 |
0.29 |
| chr3_64823974_64824125 | 0.25 |
ADAMTS9 |
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
150373 |
0.04 |
| chr1_192667639_192667790 | 0.25 |
RGS13 |
regulator of G-protein signaling 13 |
62432 |
0.13 |
| chr12_15126426_15126577 | 0.25 |
PDE6H |
phosphodiesterase 6H, cGMP-specific, cone, gamma |
545 |
0.75 |
| chr13_46927202_46927353 | 0.25 |
ENSG00000223336 |
. |
21448 |
0.17 |
| chr19_52598741_52598892 | 0.25 |
ZNF841 |
zinc finger protein 841 |
192 |
0.91 |
| chr2_143885609_143886070 | 0.25 |
ARHGAP15 |
Rho GTPase activating protein 15 |
1044 |
0.64 |
| chr20_8301190_8301355 | 0.25 |
PLCB1-IT1 |
PLCB1 intronic transcript 1 (non-protein coding) |
71900 |
0.13 |
| chr21_39811436_39811587 | 0.25 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
58834 |
0.15 |
| chr7_5764998_5765171 | 0.25 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
44992 |
0.12 |
| chr2_44978729_44978880 | 0.25 |
ENSG00000252896 |
. |
30465 |
0.24 |
| chr5_150480061_150480320 | 0.25 |
ANXA6 |
annexin A6 |
4643 |
0.22 |
| chr12_109213549_109213779 | 0.25 |
SSH1 |
slingshot protein phosphatase 1 |
6398 |
0.16 |
| chr10_7805763_7805973 | 0.25 |
KIN |
KIN, antigenic determinant of recA protein homolog (mouse) |
24076 |
0.17 |
| chr5_154350077_154350228 | 0.25 |
MRPL22 |
mitochondrial ribosomal protein L22 |
29490 |
0.14 |
| chr19_13319774_13320090 | 0.25 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
48282 |
0.08 |
| chr21_45439617_45439889 | 0.25 |
TRAPPC10 |
trafficking protein particle complex 10 |
7553 |
0.18 |
| chr13_80452469_80452769 | 0.25 |
NDFIP2 |
Nedd4 family interacting protein 2 |
397031 |
0.01 |
| chr7_142429941_142430115 | 0.24 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
27291 |
0.17 |
| chr22_38438083_38438604 | 0.24 |
RP5-1039K5.16 |
|
8108 |
0.11 |
| chr11_75585074_75585225 | 0.24 |
UVRAG |
UV radiation resistance associated |
4299 |
0.16 |
| chr17_14096680_14097352 | 0.24 |
ENSG00000252305 |
. |
16568 |
0.19 |
| chr2_113994292_113994443 | 0.24 |
ENSG00000189223 |
. |
521 |
0.65 |
| chr9_86184099_86184307 | 0.24 |
ENSG00000199483 |
. |
27296 |
0.15 |
| chr1_182182781_182183127 | 0.24 |
ENSG00000206764 |
. |
113249 |
0.06 |
| chr5_176824252_176824525 | 0.24 |
PFN3 |
profilin 3 |
3249 |
0.12 |
| chr10_74612089_74612240 | 0.24 |
OIT3 |
oncoprotein induced transcript 3 |
41175 |
0.15 |
| chr14_75807156_75807307 | 0.24 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
60335 |
0.09 |
| chr18_29636683_29636932 | 0.24 |
ENSG00000265063 |
. |
4145 |
0.17 |
| chr1_207243907_207244203 | 0.24 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
5706 |
0.14 |
| chr12_11874823_11874974 | 0.24 |
ETV6 |
ets variant 6 |
30537 |
0.23 |
| chr7_141565022_141565323 | 0.24 |
OR9A3P |
olfactory receptor, family 9, subfamily A, member 3 pseudogene |
2512 |
0.17 |
| chr11_10836352_10836503 | 0.24 |
RP11-685M7.3 |
|
5583 |
0.14 |
| chrX_65125867_65126018 | 0.24 |
RP6-159A1.3 |
|
93651 |
0.08 |
| chr14_91844279_91844530 | 0.24 |
CCDC88C |
coiled-coil domain containing 88C |
39286 |
0.15 |
| chr15_77707627_77707803 | 0.24 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
4727 |
0.25 |
| chr18_24281825_24282055 | 0.24 |
ENSG00000265369 |
. |
12447 |
0.22 |
| chr12_71401978_71402129 | 0.23 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
87430 |
0.08 |
| chr17_14233058_14233209 | 0.23 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
28733 |
0.23 |
| chr13_72400590_72400741 | 0.23 |
DACH1 |
dachshund homolog 1 (Drosophila) |
40242 |
0.22 |
| chr1_79111038_79111189 | 0.23 |
IFI44 |
interferon-induced protein 44 |
4403 |
0.25 |
| chr1_207095862_207096179 | 0.23 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
521 |
0.73 |
| chr16_66619782_66620057 | 0.23 |
RP11-403P17.2 |
|
5300 |
0.11 |
| chr7_37706259_37706410 | 0.23 |
GPR141 |
G protein-coupled receptor 141 |
17066 |
0.22 |
| chr21_45774588_45774760 | 0.23 |
TRPM2 |
transient receptor potential cation channel, subfamily M, member 2 |
1103 |
0.32 |
| chr10_64394826_64394983 | 0.23 |
ZNF365 |
zinc finger protein 365 |
8629 |
0.24 |
| chr12_104894058_104894320 | 0.23 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
43410 |
0.18 |
| chrX_135064888_135065039 | 0.23 |
SLC9A6 |
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6 |
2635 |
0.27 |
| chr3_196322790_196323004 | 0.23 |
FBXO45 |
F-box protein 45 |
27338 |
0.1 |
| chr18_54135220_54135371 | 0.23 |
TXNL1 |
thioredoxin-like 1 |
146427 |
0.05 |
| chr2_198099581_198099897 | 0.23 |
ANKRD44 |
ankyrin repeat domain 44 |
36977 |
0.14 |
| chr9_91927372_91927649 | 0.23 |
ENSG00000265112 |
. |
370 |
0.8 |
| chr7_20510108_20510347 | 0.23 |
ENSG00000200753 |
. |
93273 |
0.08 |
| chr6_139969651_139969888 | 0.23 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
274012 |
0.02 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.2 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.3 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.6 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.0 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.1 | GO:1903020 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.0 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.4 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.2 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.1 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.1 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.0 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.0 | GO:0002604 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.0 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0036230 | granulocyte activation(GO:0036230) |
| 0.0 | 0.1 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.0 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.0 | GO:1903224 | endodermal cell fate commitment(GO:0001711) endodermal cell fate specification(GO:0001714) endodermal cell differentiation(GO:0035987) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.3 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0042685 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.0 | GO:0044144 | growth involved in symbiotic interaction(GO:0044110) growth of symbiont involved in interaction with host(GO:0044116) growth of symbiont in host(GO:0044117) regulation of growth of symbiont in host(GO:0044126) negative regulation of growth of symbiont in host(GO:0044130) modulation of growth of symbiont involved in interaction with host(GO:0044144) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.0 | GO:0010613 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 | 0.1 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.1 | GO:0009595 | detection of biotic stimulus(GO:0009595) |
| 0.0 | 0.0 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.0 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0010829 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0060346 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.1 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.0 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0051657 | maintenance of organelle location(GO:0051657) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0031083 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.0 | GO:0001520 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0043073 | germ cell nucleus(GO:0043073) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.4 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0032357 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.3 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.0 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.0 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |