Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
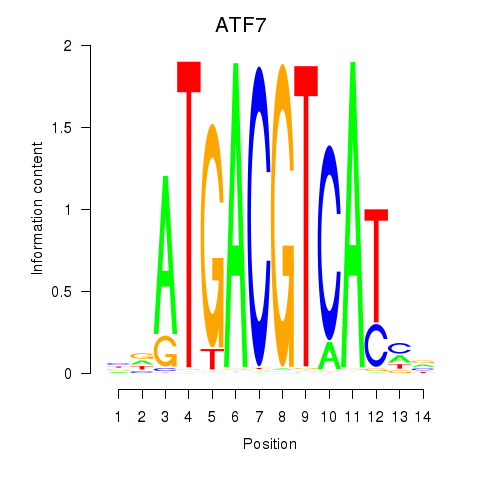
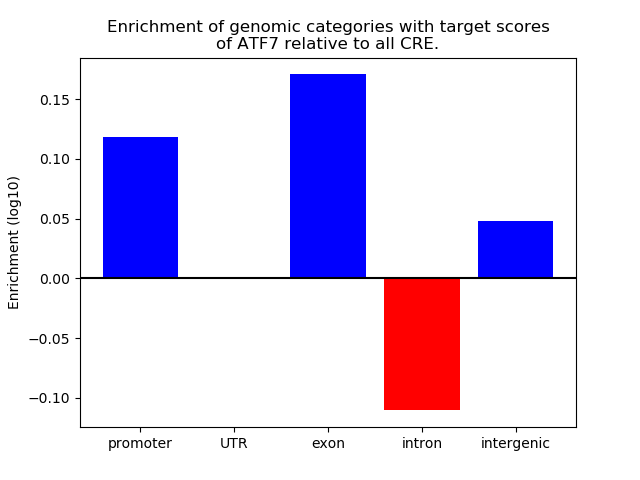
Results for ATF7
Z-value: 0.90
Transcription factors associated with ATF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF7
|
ENSG00000170653.14 | activating transcription factor 7 |
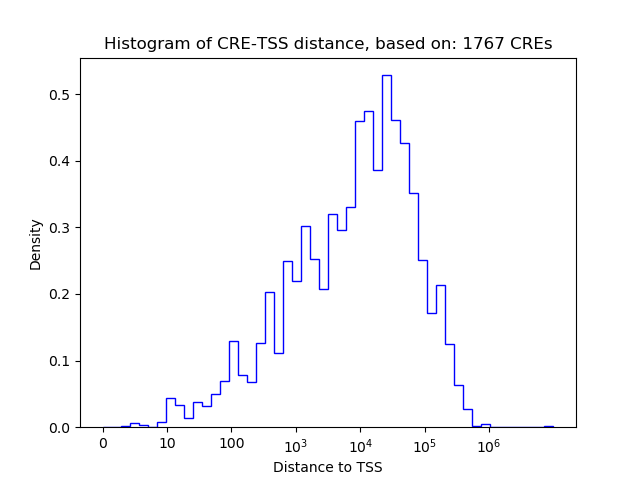
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_53964156_53964307 | ATF7 | 30574 | 0.109163 | 0.74 | 2.2e-02 | Click! |
| chr12_53989142_53989293 | ATF7 | 5588 | 0.168089 | 0.74 | 2.3e-02 | Click! |
| chr12_54017496_54017647 | ATF7 | 2184 | 0.226720 | 0.74 | 2.4e-02 | Click! |
| chr12_53969438_53969764 | ATF7 | 25204 | 0.121145 | 0.72 | 2.9e-02 | Click! |
| chr12_53969924_53970134 | ATF7 | 24776 | 0.122080 | 0.71 | 3.2e-02 | Click! |
Activity of the ATF7 motif across conditions
Conditions sorted by the z-value of the ATF7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
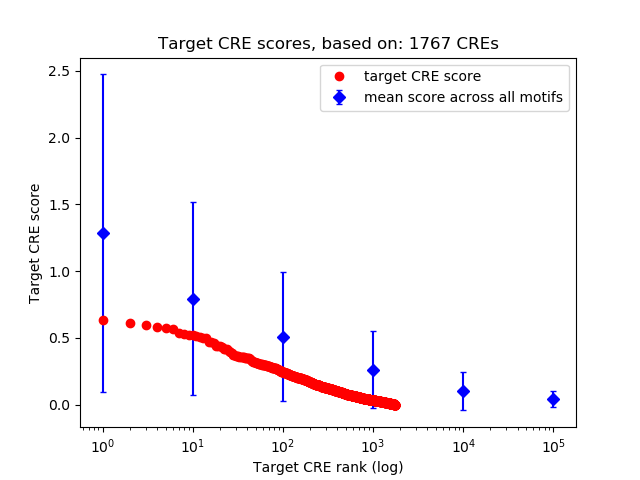
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_141015090_141015375 | 0.64 |
RP11-392B6.1 |
|
33937 |
0.2 |
| chr6_80714655_80715350 | 0.61 |
TTK |
TTK protein kinase |
451 |
0.88 |
| chr5_14264414_14265476 | 0.60 |
TRIO |
trio Rho guanine nucleotide exchange factor |
26141 |
0.27 |
| chr1_56981894_56982239 | 0.59 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
63175 |
0.14 |
| chr12_133022629_133022780 | 0.57 |
MUC8 |
mucin 8 |
28022 |
0.17 |
| chr2_8552253_8552511 | 0.57 |
AC011747.7 |
|
263514 |
0.02 |
| chr5_73730230_73730381 | 0.53 |
ENSG00000244326 |
. |
116494 |
0.06 |
| chr15_40636361_40636512 | 0.53 |
C15orf52 |
chromosome 15 open reading frame 52 |
3268 |
0.1 |
| chr11_12695755_12695947 | 0.52 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
118 |
0.98 |
| chr6_64516560_64517136 | 0.52 |
RP11-59D5__B.2 |
|
119 |
0.98 |
| chr6_14477462_14477818 | 0.51 |
ENSG00000206960 |
. |
169126 |
0.04 |
| chr17_65505786_65506259 | 0.51 |
CTD-2653B5.1 |
|
14575 |
0.17 |
| chr21_44937001_44937451 | 0.50 |
SIK1 |
salt-inducible kinase 1 |
90218 |
0.08 |
| chr16_31141784_31142119 | 0.50 |
RP11-388M20.2 |
|
803 |
0.33 |
| chr12_8043408_8043946 | 0.47 |
SLC2A14 |
solute carrier family 2 (facilitated glucose transporter), member 14 |
67 |
0.95 |
| chr6_2449367_2449525 | 0.47 |
ENSG00000266252 |
. |
39577 |
0.21 |
| chr1_181056660_181057120 | 0.46 |
IER5 |
immediate early response 5 |
748 |
0.71 |
| chr4_74575921_74576072 | 0.44 |
IL8 |
interleukin 8 |
30227 |
0.19 |
| chr5_179453537_179453707 | 0.44 |
ENSG00000198995 |
. |
11225 |
0.19 |
| chr14_75655646_75655840 | 0.44 |
TMED10 |
transmembrane emp24-like trafficking protein 10 (yeast) |
12409 |
0.15 |
| chr7_100782348_100782499 | 0.44 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
12044 |
0.1 |
| chr10_6959147_6959298 | 0.42 |
PRKCQ |
protein kinase C, theta |
336959 |
0.01 |
| chr1_17630639_17630914 | 0.42 |
PADI4 |
peptidyl arginine deiminase, type IV |
3914 |
0.2 |
| chr9_130912677_130912874 | 0.41 |
LCN2 |
lipocalin 2 |
978 |
0.34 |
| chr1_22380057_22380208 | 0.40 |
CDC42 |
cell division cycle 42 |
342 |
0.82 |
| chr22_44569121_44569281 | 0.39 |
PARVG |
parvin, gamma |
365 |
0.92 |
| chr8_62625004_62625155 | 0.39 |
ASPH |
aspartate beta-hydroxylase |
2005 |
0.33 |
| chr11_65256298_65256486 | 0.38 |
AP000769.1 |
Uncharacterized protein |
33664 |
0.08 |
| chr4_153913148_153913299 | 0.37 |
FHDC1 |
FH2 domain containing 1 |
49088 |
0.16 |
| chr8_61896258_61896409 | 0.36 |
CLVS1 |
clavesin 1 |
73384 |
0.12 |
| chr17_25773754_25774039 | 0.36 |
KSR1 |
kinase suppressor of ras 1 |
9774 |
0.19 |
| chr2_28114153_28114304 | 0.36 |
RBKS |
ribokinase |
263 |
0.74 |
| chr15_49336349_49337349 | 0.36 |
SECISBP2L |
SECIS binding protein 2-like |
1781 |
0.28 |
| chr6_18403082_18403233 | 0.36 |
ENSG00000238458 |
. |
874 |
0.65 |
| chr17_55446479_55446630 | 0.36 |
ENSG00000263902 |
. |
43008 |
0.17 |
| chr17_42423914_42424082 | 0.36 |
GRN |
granulin |
1325 |
0.25 |
| chr3_99721724_99721891 | 0.36 |
ENSG00000264897 |
. |
38565 |
0.19 |
| chr2_108994281_108994432 | 0.35 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
112 |
0.97 |
| chr9_111578900_111579091 | 0.35 |
ACTL7B |
actin-like 7B |
40244 |
0.16 |
| chr18_10791557_10791762 | 0.35 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
11 |
0.99 |
| chr17_61723267_61723570 | 0.35 |
MAP3K3 |
mitogen-activated protein kinase kinase kinase 3 |
23348 |
0.13 |
| chr12_6187508_6187659 | 0.35 |
ENSG00000240533 |
. |
17553 |
0.21 |
| chr10_11628107_11628334 | 0.34 |
USP6NL |
USP6 N-terminal like |
12380 |
0.23 |
| chr2_197944933_197945147 | 0.33 |
ANKRD44 |
ankyrin repeat domain 44 |
30528 |
0.22 |
| chr17_42978313_42978555 | 0.33 |
CCDC103 |
coiled-coil domain containing 103 |
1292 |
0.19 |
| chr13_99234265_99234566 | 0.32 |
STK24-AS1 |
STK24 antisense RNA 1 |
4917 |
0.23 |
| chr13_97876107_97876342 | 0.32 |
MBNL2 |
muscleblind-like splicing regulator 2 |
1615 |
0.53 |
| chr8_48393092_48393308 | 0.32 |
SPIDR |
scaffolding protein involved in DNA repair |
40237 |
0.18 |
| chr2_8144515_8144736 | 0.32 |
ENSG00000221255 |
. |
427653 |
0.01 |
| chr18_20840672_20840823 | 0.31 |
RP11-17J14.2 |
|
429 |
0.89 |
| chr20_36797041_36797355 | 0.31 |
TGM2 |
transglutaminase 2 |
2218 |
0.32 |
| chr8_49838453_49838744 | 0.31 |
SNAI2 |
snail family zinc finger 2 |
4299 |
0.35 |
| chr6_53213778_53213929 | 0.31 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
48 |
0.98 |
| chr16_70730831_70731276 | 0.31 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
1557 |
0.31 |
| chr5_102729544_102729916 | 0.31 |
C5orf30 |
chromosome 5 open reading frame 30 |
134605 |
0.05 |
| chr7_132095239_132095390 | 0.30 |
AC011625.1 |
|
58221 |
0.15 |
| chr10_103143872_103144023 | 0.30 |
ENSG00000239091 |
. |
12238 |
0.16 |
| chr16_73087544_73087838 | 0.30 |
ZFHX3 |
zinc finger homeobox 3 |
5417 |
0.25 |
| chr1_113361578_113361925 | 0.30 |
FAM19A3 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A3 |
98552 |
0.05 |
| chr4_38822000_38822151 | 0.30 |
TLR6 |
toll-like receptor 6 |
9085 |
0.16 |
| chr14_73927648_73927878 | 0.30 |
ENSG00000251393 |
. |
1366 |
0.34 |
| chr12_124983629_124983780 | 0.30 |
NCOR2 |
nuclear receptor corepressor 2 |
3906 |
0.36 |
| chr17_18528921_18529072 | 0.30 |
CCDC144B |
coiled-coil domain containing 144B (pseudogene) |
75 |
0.96 |
| chr11_13238293_13238470 | 0.30 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
59818 |
0.15 |
| chr4_159131101_159131306 | 0.29 |
TMEM144 |
transmembrane protein 144 |
143 |
0.96 |
| chr18_22820121_22820315 | 0.29 |
ZNF521 |
zinc finger protein 521 |
8724 |
0.3 |
| chr15_91040083_91040234 | 0.29 |
CRTC3 |
CREB regulated transcription coactivator 3 |
32999 |
0.15 |
| chr3_159571102_159571298 | 0.29 |
SCHIP1 |
schwannomin interacting protein 1 |
472 |
0.82 |
| chr4_15721261_15721412 | 0.29 |
BST1 |
bone marrow stromal cell antigen 1 |
4385 |
0.23 |
| chr8_74792715_74792866 | 0.29 |
UBE2W |
ubiquitin-conjugating enzyme E2W (putative) |
1645 |
0.41 |
| chr6_134803152_134803303 | 0.29 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
163977 |
0.04 |
| chr3_105178540_105178691 | 0.29 |
ALCAM |
activated leukocyte cell adhesion molecule |
53554 |
0.19 |
| chr4_174198585_174198817 | 0.28 |
ENSG00000221296 |
. |
9390 |
0.17 |
| chr3_124500151_124500305 | 0.28 |
ITGB5-AS1 |
ITGB5 antisense RNA 1 |
226 |
0.93 |
| chr15_94850174_94850340 | 0.28 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
8762 |
0.33 |
| chr2_102423827_102423978 | 0.28 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
10176 |
0.28 |
| chr2_65309092_65309434 | 0.28 |
RAB1A |
RAB1A, member RAS oncogene family |
24261 |
0.15 |
| chr9_137348457_137348644 | 0.28 |
RXRA |
retinoid X receptor, alpha |
50122 |
0.15 |
| chr1_11901364_11901515 | 0.28 |
NPPA-AS1 |
NPPA antisense RNA 1 |
365 |
0.78 |
| chr3_10215716_10215867 | 0.27 |
IRAK2 |
interleukin-1 receptor-associated kinase 2 |
9242 |
0.12 |
| chr1_199147885_199148036 | 0.27 |
ENSG00000239006 |
. |
18352 |
0.3 |
| chr7_6183718_6183869 | 0.27 |
USP42 |
ubiquitin specific peptidase 42 |
27682 |
0.11 |
| chr8_66761852_66762003 | 0.27 |
PDE7A |
phosphodiesterase 7A |
7740 |
0.3 |
| chr17_19206898_19207138 | 0.27 |
EPN2-AS1 |
EPN2 antisense RNA 1 |
2556 |
0.18 |
| chr12_108958034_108958185 | 0.27 |
ISCU |
iron-sulfur cluster assembly enzyme |
1723 |
0.27 |
| chr2_38762873_38763024 | 0.27 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
34030 |
0.18 |
| chrX_18665752_18666019 | 0.27 |
RS1 |
retinoschisin 1 |
24344 |
0.19 |
| chr20_45934513_45934664 | 0.26 |
ENSG00000265532 |
. |
2538 |
0.2 |
| chr1_161053172_161053323 | 0.26 |
RP11-544M22.8 |
|
1008 |
0.29 |
| chr16_86817580_86817731 | 0.26 |
FOXL1 |
forkhead box L1 |
205540 |
0.02 |
| chr9_139117528_139117679 | 0.26 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
880 |
0.58 |
| chr10_121580538_121580689 | 0.26 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
2385 |
0.32 |
| chr10_134826309_134826460 | 0.25 |
GPR123 |
G protein-coupled receptor 123 |
58049 |
0.11 |
| chr7_12588835_12588986 | 0.25 |
SCIN |
scinderin |
21402 |
0.21 |
| chr13_29156883_29157128 | 0.25 |
POMP |
proteasome maturation protein |
76236 |
0.1 |
| chr9_114288073_114288445 | 0.25 |
ZNF483 |
zinc finger protein 483 |
746 |
0.65 |
| chr14_68608308_68608459 | 0.25 |
ENSG00000244677 |
. |
8242 |
0.23 |
| chr16_70456809_70457112 | 0.25 |
ST3GAL2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
16031 |
0.11 |
| chr1_48709961_48710211 | 0.24 |
RP5-1024N4.4 |
|
14853 |
0.22 |
| chr9_95895847_95895998 | 0.24 |
NINJ1 |
ninjurin 1 |
648 |
0.7 |
| chr9_91390340_91390723 | 0.24 |
ENSG00000265873 |
. |
29711 |
0.24 |
| chr7_26726297_26726654 | 0.24 |
C7orf71 |
chromosome 7 open reading frame 71 |
48985 |
0.17 |
| chr19_32837185_32838232 | 0.24 |
ZNF507 |
zinc finger protein 507 |
1083 |
0.51 |
| chr1_186445670_186446022 | 0.24 |
PDC |
phosducin |
15592 |
0.19 |
| chr17_7369936_7370087 | 0.24 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
11098 |
0.06 |
| chr5_134726191_134726464 | 0.24 |
H2AFY |
H2A histone family, member Y |
8574 |
0.16 |
| chr15_43531002_43531179 | 0.24 |
ENSG00000202211 |
. |
8788 |
0.13 |
| chr8_126374976_126375409 | 0.24 |
RP11-550A5.2 |
|
11534 |
0.24 |
| chr12_66429713_66429864 | 0.24 |
ENSG00000251857 |
. |
30213 |
0.17 |
| chr19_996464_996712 | 0.24 |
AC004528.1 |
Uncharacterized protein |
3013 |
0.1 |
| chr8_65490045_65490217 | 0.23 |
RP11-21C4.1 |
|
311 |
0.91 |
| chr1_28099274_28099549 | 0.23 |
STX12 |
syntaxin 12 |
283 |
0.78 |
| chr6_11207713_11207934 | 0.23 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
25068 |
0.18 |
| chr10_120291049_120291200 | 0.23 |
ENSG00000266281 |
. |
14482 |
0.25 |
| chr10_35776302_35776593 | 0.23 |
GJD4 |
gap junction protein, delta 4, 40.1kDa |
117891 |
0.05 |
| chr3_37366423_37366574 | 0.23 |
ENSG00000222208 |
. |
16642 |
0.18 |
| chr2_45464381_45464651 | 0.23 |
SIX2 |
SIX homeobox 2 |
227947 |
0.02 |
| chr11_70270544_70270893 | 0.23 |
CTTN |
cortactin |
4118 |
0.18 |
| chr5_172222413_172222569 | 0.23 |
ENSG00000206741 |
. |
15394 |
0.14 |
| chr1_205216416_205216567 | 0.23 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
743 |
0.63 |
| chr7_138914973_138915166 | 0.22 |
UBN2 |
ubinuclein 2 |
33 |
0.98 |
| chr15_58653715_58653866 | 0.22 |
LIPC |
lipase, hepatic |
48978 |
0.15 |
| chr9_135115291_135115736 | 0.22 |
ENSG00000221105 |
. |
25251 |
0.2 |
| chr6_41680623_41680774 | 0.22 |
TFEB |
transcription factor EB |
7020 |
0.13 |
| chr3_70903414_70903565 | 0.22 |
ENSG00000221382 |
. |
24454 |
0.23 |
| chr19_35849271_35849508 | 0.22 |
FFAR3 |
free fatty acid receptor 3 |
27 |
0.95 |
| chr19_8454744_8454908 | 0.22 |
RAB11B |
RAB11B, member RAS oncogene family |
39 |
0.64 |
| chr20_62580456_62580607 | 0.22 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
1948 |
0.13 |
| chr21_40386888_40387293 | 0.21 |
ENSG00000272015 |
. |
120381 |
0.05 |
| chr19_11402892_11403043 | 0.21 |
TSPAN16 |
tetraspanin 16 |
3857 |
0.12 |
| chr5_169918557_169918708 | 0.21 |
CTB-147C13.1 |
|
5221 |
0.24 |
| chr4_139156572_139156855 | 0.21 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
6790 |
0.31 |
| chr4_8263775_8264072 | 0.21 |
HTRA3 |
HtrA serine peptidase 3 |
7569 |
0.22 |
| chr3_99679872_99680023 | 0.21 |
ENSG00000264897 |
. |
3295 |
0.35 |
| chr12_54623516_54623843 | 0.21 |
ENSG00000265371 |
. |
1581 |
0.23 |
| chr9_78990142_78990293 | 0.21 |
RFK |
riboflavin kinase |
18831 |
0.24 |
| chr4_78783004_78783324 | 0.21 |
MRPL1 |
mitochondrial ribosomal protein L1 |
510 |
0.86 |
| chr4_157114740_157114891 | 0.21 |
ENSG00000221189 |
. |
102983 |
0.08 |
| chr10_50452094_50452371 | 0.21 |
C10orf71-AS1 |
C10orf71 antisense RNA 1 |
54593 |
0.1 |
| chr20_35110278_35110568 | 0.21 |
DLGAP4 |
discs, large (Drosophila) homolog-associated protein 4 |
20273 |
0.17 |
| chr12_62861008_62861159 | 0.21 |
MON2 |
MON2 homolog (S. cerevisiae) |
454 |
0.83 |
| chr11_116766141_116766351 | 0.21 |
SIK3-IT1 |
SIK3 intronic transcript 1 (non-protein coding) |
4809 |
0.17 |
| chr8_26465978_26466617 | 0.20 |
DPYSL2 |
dihydropyrimidinase-like 2 |
30376 |
0.22 |
| chr6_166879262_166879723 | 0.20 |
RPS6KA2-IT1 |
RPS6KA2 intronic transcript 1 (non-protein coding) |
621 |
0.74 |
| chr4_8430147_8430298 | 0.20 |
ACOX3 |
acyl-CoA oxidase 3, pristanoyl |
9 |
0.98 |
| chr1_152005862_152006666 | 0.20 |
S100A11 |
S100 calcium binding protein A11 |
3247 |
0.19 |
| chr12_54656131_54656480 | 0.20 |
RP11-968A15.2 |
|
94 |
0.92 |
| chr18_60770372_60770523 | 0.20 |
RP11-299P2.1 |
|
48106 |
0.16 |
| chr5_148130748_148130917 | 0.20 |
ADRB2 |
adrenoceptor beta 2, surface |
75324 |
0.1 |
| chr5_82771804_82772197 | 0.20 |
VCAN |
versican |
4256 |
0.33 |
| chr9_124091436_124091643 | 0.20 |
GSN |
gelsolin |
2679 |
0.21 |
| chr17_34115756_34115930 | 0.20 |
MMP28 |
matrix metallopeptidase 28 |
6868 |
0.1 |
| chr5_64331605_64331756 | 0.20 |
ENSG00000207439 |
. |
87516 |
0.1 |
| chr12_124997656_124997821 | 0.20 |
NCOR2 |
nuclear receptor corepressor 2 |
5102 |
0.34 |
| chr17_60970180_60970441 | 0.20 |
ENSG00000207552 |
. |
51266 |
0.14 |
| chr1_151254019_151254394 | 0.20 |
ZNF687 |
zinc finger protein 687 |
112 |
0.61 |
| chr5_126111660_126112314 | 0.20 |
LMNB1 |
lamin B1 |
853 |
0.67 |
| chr6_15463967_15464118 | 0.20 |
JARID2 |
jumonji, AT rich interactive domain 2 |
62953 |
0.13 |
| chr13_113664073_113664237 | 0.20 |
RP11-120K24.5 |
|
170 |
0.9 |
| chr6_21925832_21925983 | 0.20 |
ENSG00000222515 |
. |
159866 |
0.04 |
| chr19_18491287_18491451 | 0.20 |
GDF15 |
growth differentiation factor 15 |
1611 |
0.2 |
| chr16_58535345_58535501 | 0.20 |
NDRG4 |
NDRG family member 4 |
16 |
0.97 |
| chr12_27939071_27939324 | 0.20 |
RP11-860B13.1 |
|
5197 |
0.13 |
| chrY_2576678_2576907 | 0.20 |
ENSG00000251841 |
. |
75998 |
0.11 |
| chr7_993508_993659 | 0.19 |
ADAP1 |
ArfGAP with dual PH domains 1 |
750 |
0.56 |
| chr13_72430784_72430935 | 0.19 |
DACH1 |
dachshund homolog 1 (Drosophila) |
10048 |
0.32 |
| chr9_139117754_139118099 | 0.19 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
1203 |
0.46 |
| chr2_102093131_102093315 | 0.19 |
RFX8 |
RFX family member 8, lacking RFX DNA binding domain |
2058 |
0.43 |
| chr12_94625674_94625825 | 0.19 |
PLXNC1 |
plexin C1 |
22545 |
0.17 |
| chr17_53573843_53573994 | 0.19 |
ENSG00000200107 |
. |
55253 |
0.15 |
| chr5_7803203_7803354 | 0.19 |
C5orf49 |
chromosome 5 open reading frame 49 |
47986 |
0.13 |
| chrX_110339162_110339436 | 0.19 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
289 |
0.95 |
| chr8_41625601_41625752 | 0.19 |
ANK1 |
ankyrin 1, erythrocytic |
29464 |
0.15 |
| chr1_151508284_151508566 | 0.19 |
TUFT1 |
tuftelin 1 |
4356 |
0.12 |
| chr5_177900691_177901194 | 0.19 |
CTB-26E19.1 |
|
30847 |
0.19 |
| chr3_171592714_171592972 | 0.19 |
TMEM212-IT1 |
TMEM212 intronic transcript 1 (non-protein coding) |
19383 |
0.19 |
| chr7_132217204_132217355 | 0.19 |
PLXNA4 |
plexin A4 |
43950 |
0.19 |
| chr13_107137040_107137296 | 0.19 |
EFNB2 |
ephrin-B2 |
50294 |
0.17 |
| chr1_223724894_223725187 | 0.19 |
CAPN8 |
calpain 8 |
660 |
0.81 |
| chr8_101348735_101348886 | 0.18 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
378 |
0.89 |
| chr13_80444406_80444557 | 0.18 |
NDFIP2 |
Nedd4 family interacting protein 2 |
388893 |
0.01 |
| chr6_113075862_113076013 | 0.18 |
ENSG00000201386 |
. |
216758 |
0.02 |
| chr15_52138768_52139605 | 0.18 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
15849 |
0.15 |
| chr17_9273783_9274123 | 0.18 |
AC087501.1 |
|
81561 |
0.08 |
| chr3_32451109_32451284 | 0.18 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
17665 |
0.23 |
| chr12_89511566_89511717 | 0.18 |
ENSG00000238302 |
. |
164421 |
0.04 |
| chr7_5258466_5258734 | 0.18 |
WIPI2 |
WD repeat domain, phosphoinositide interacting 2 |
4700 |
0.2 |
| chr7_20341021_20341275 | 0.18 |
ITGB8 |
integrin, beta 8 |
29177 |
0.17 |
| chr17_57971307_57971993 | 0.18 |
RPS6KB1 |
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
1050 |
0.37 |
| chr9_111034278_111034617 | 0.18 |
ENSG00000222512 |
. |
86762 |
0.11 |
| chr1_43941761_43941966 | 0.18 |
HYI |
hydroxypyruvate isomerase (putative) |
22203 |
0.12 |
| chr6_34992306_34992644 | 0.18 |
TCP11 |
t-complex 11, testis-specific |
96347 |
0.07 |
| chr3_30668319_30669005 | 0.17 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
20569 |
0.26 |
| chr3_172345425_172345576 | 0.17 |
AC007919.2 |
HCG1787166; PRO1163; Uncharacterized protein |
15983 |
0.19 |
| chr9_134516940_134517091 | 0.17 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
15680 |
0.22 |
| chr18_33613590_33613806 | 0.17 |
RPRD1A |
regulation of nuclear pre-mRNA domain containing 1A |
2885 |
0.3 |
| chr2_39721987_39722138 | 0.17 |
AC007246.3 |
|
19225 |
0.22 |
| chr2_233367753_233368028 | 0.17 |
ECEL1 |
endothelin converting enzyme-like 1 |
15352 |
0.1 |
| chr9_139810950_139811101 | 0.17 |
RP11-229P13.25 |
|
21415 |
0.06 |
| chr10_44506456_44506924 | 0.17 |
ENSG00000238957 |
. |
23298 |
0.27 |
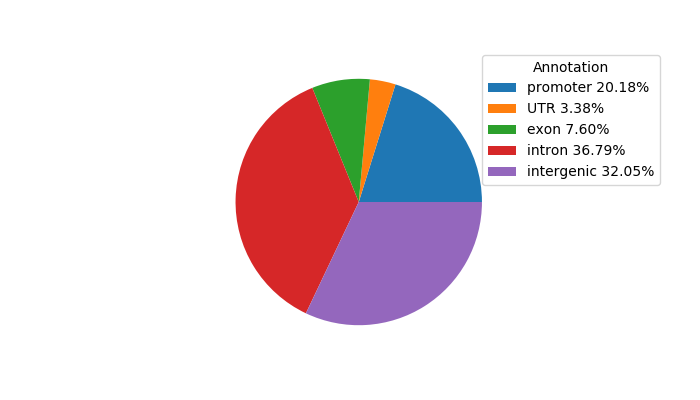
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.3 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0010523 | negative regulation of calcium ion transport into cytosol(GO:0010523) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.2 | GO:0071347 | cellular response to interleukin-1(GO:0071347) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0072191 | positive regulation of smooth muscle cell differentiation(GO:0051152) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |