Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
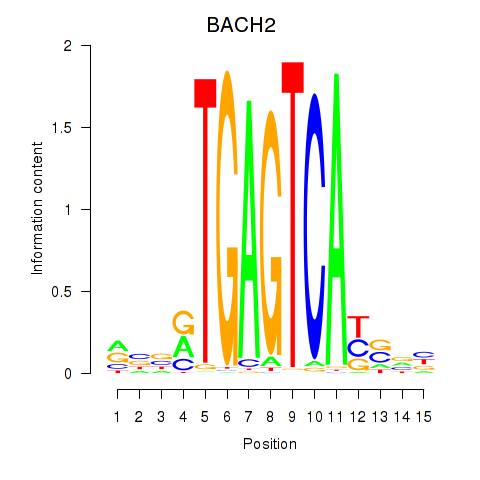
Results for BACH2
Z-value: 0.74
Transcription factors associated with BACH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH2
|
ENSG00000112182.10 | BTB domain and CNC homolog 2 |
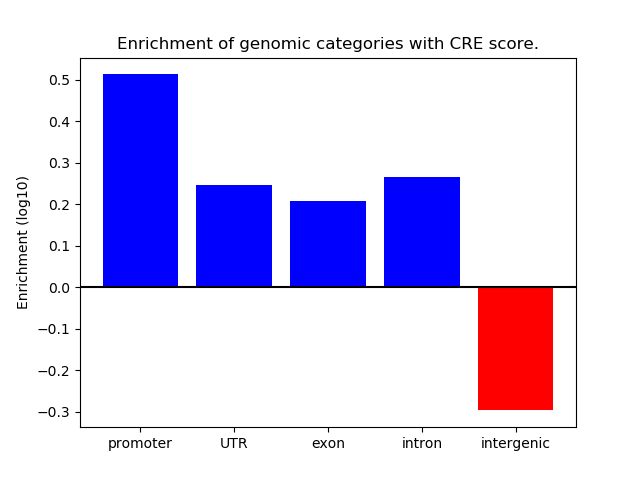
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_90981476_90981674 | BACH2 | 24886 | 0.218364 | -0.90 | 1.0e-03 | Click! |
| chr6_90981736_90981993 | BACH2 | 24597 | 0.218974 | -0.89 | 1.3e-03 | Click! |
| chr6_90982113_90982279 | BACH2 | 24265 | 0.219669 | -0.86 | 2.6e-03 | Click! |
| chr6_90984432_90984583 | BACH2 | 21954 | 0.224353 | -0.83 | 5.1e-03 | Click! |
| chr6_90951264_90951456 | BACH2 | 55101 | 0.146848 | -0.82 | 6.5e-03 | Click! |
Activity of the BACH2 motif across conditions
Conditions sorted by the z-value of the BACH2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
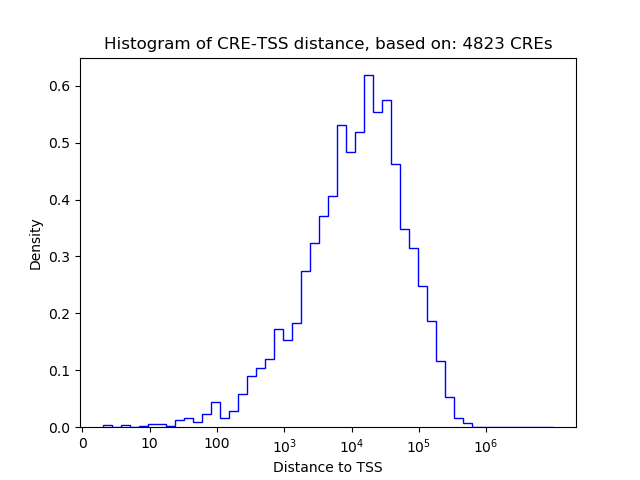
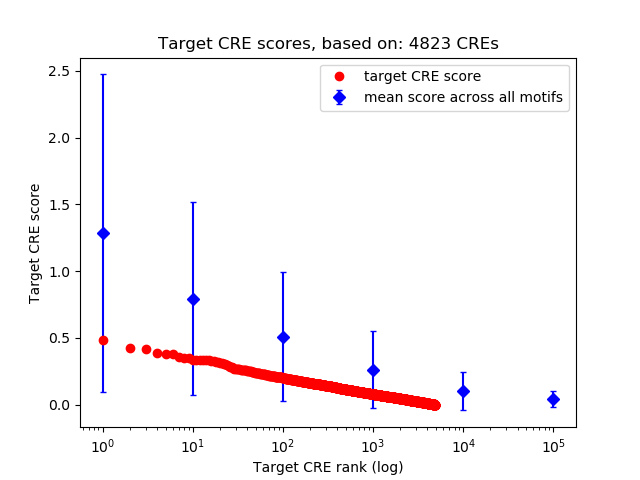
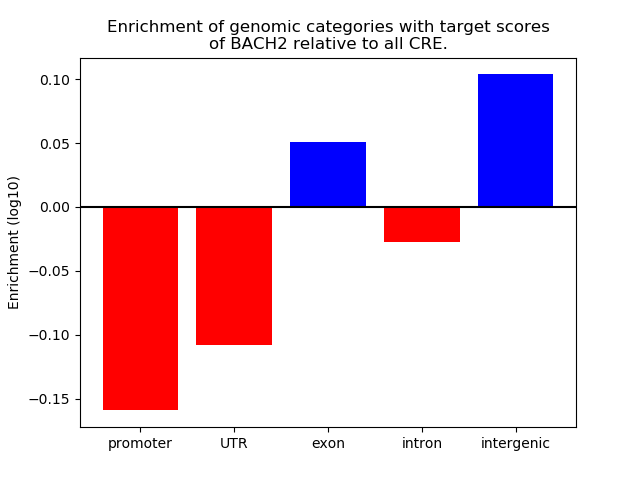
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_18539696_18539847 | 0.49 |
XXbac-B476C20.9 |
|
20792 |
0.13 |
| chr2_231481752_231482027 | 0.42 |
ENSG00000199791 |
. |
30053 |
0.17 |
| chr1_232845212_232845363 | 0.42 |
ENSG00000238382 |
. |
9337 |
0.28 |
| chr2_128155940_128156196 | 0.39 |
MAP3K2 |
mitogen-activated protein kinase kinase kinase 2 |
10027 |
0.16 |
| chr2_110316196_110316465 | 0.38 |
AC011753.5 |
|
46945 |
0.13 |
| chr4_8192745_8193409 | 0.38 |
SH3TC1 |
SH3 domain and tetratricopeptide repeats 1 |
8014 |
0.22 |
| chr15_52393088_52393274 | 0.36 |
CTD-2184D3.5 |
|
461 |
0.77 |
| chr2_43303091_43303242 | 0.35 |
ENSG00000207087 |
. |
15466 |
0.28 |
| chr19_39224351_39224502 | 0.35 |
CAPN12 |
calpain 12 |
1619 |
0.22 |
| chr12_66089171_66089322 | 0.34 |
HMGA2 |
high mobility group AT-hook 2 |
128665 |
0.05 |
| chr12_6388900_6389082 | 0.34 |
PLEKHG6 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
30611 |
0.12 |
| chr8_145993953_145994104 | 0.34 |
ZNF34 |
zinc finger protein 34 |
11639 |
0.11 |
| chr2_26235492_26235765 | 0.33 |
AC013449.1 |
Uncharacterized protein |
15853 |
0.16 |
| chr8_102038362_102038931 | 0.33 |
ENSG00000252736 |
. |
63375 |
0.1 |
| chr4_144276820_144277057 | 0.33 |
ENSG00000265623 |
. |
12325 |
0.2 |
| chr2_225544034_225544185 | 0.33 |
CUL3 |
cullin 3 |
93999 |
0.09 |
| chr17_40561664_40561962 | 0.33 |
ENSG00000221020 |
. |
10786 |
0.11 |
| chr1_244437634_244437902 | 0.32 |
C1orf100 |
chromosome 1 open reading frame 100 |
78169 |
0.1 |
| chr6_41680801_41681561 | 0.32 |
RP11-298J23.5 |
|
6953 |
0.13 |
| chr12_56120404_56120555 | 0.32 |
CD63 |
CD63 molecule |
364 |
0.7 |
| chr4_10097090_10097875 | 0.31 |
ENSG00000264931 |
. |
17166 |
0.14 |
| chr11_58395317_58395468 | 0.30 |
CNTF |
ciliary neurotrophic factor |
5246 |
0.17 |
| chr3_112500584_112500735 | 0.30 |
ENSG00000242770 |
. |
20666 |
0.22 |
| chr17_48128276_48128773 | 0.29 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
4815 |
0.16 |
| chr19_16175455_16175640 | 0.29 |
TPM4 |
tropomyosin 4 |
2284 |
0.28 |
| chr15_74688962_74689487 | 0.28 |
CYP11A1 |
cytochrome P450, family 11, subfamily A, polypeptide 1 |
29143 |
0.11 |
| chr17_75118639_75118790 | 0.28 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
5241 |
0.22 |
| chr13_114524672_114524969 | 0.27 |
GAS6-AS1 |
GAS6 antisense RNA 1 |
6217 |
0.22 |
| chr1_31229795_31230155 | 0.27 |
LAPTM5 |
lysosomal protein transmembrane 5 |
692 |
0.65 |
| chr14_59827605_59828209 | 0.27 |
ENSG00000252869 |
. |
38834 |
0.18 |
| chr3_119854481_119854726 | 0.27 |
ENSG00000244139 |
. |
13847 |
0.18 |
| chr14_62052476_62052660 | 0.27 |
RP11-47I22.3 |
Uncharacterized protein |
15254 |
0.21 |
| chr15_74281262_74281725 | 0.27 |
STOML1 |
stomatin (EPB72)-like 1 |
2682 |
0.22 |
| chr5_168521975_168522126 | 0.26 |
CTB-174D11.1 |
|
64607 |
0.13 |
| chr13_33836245_33836396 | 0.26 |
ENSG00000236581 |
. |
9196 |
0.23 |
| chr1_1240872_1241150 | 0.26 |
ACAP3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
2258 |
0.11 |
| chr14_59895317_59895579 | 0.26 |
ENSG00000252869 |
. |
28707 |
0.16 |
| chr9_124044825_124044976 | 0.26 |
RP11-477J21.6 |
|
108 |
0.95 |
| chr1_10519294_10519577 | 0.26 |
RP5-1113E3.3 |
|
823 |
0.46 |
| chr16_70780334_70780534 | 0.26 |
RP11-394B2.6 |
|
526 |
0.69 |
| chr5_148865258_148865409 | 0.25 |
CSNK1A1 |
casein kinase 1, alpha 1 |
27387 |
0.14 |
| chr8_41655772_41655923 | 0.25 |
ANK1 |
ankyrin 1, erythrocytic |
707 |
0.67 |
| chr9_130326042_130326221 | 0.25 |
FAM129B |
family with sequence similarity 129, member B |
5236 |
0.19 |
| chr17_29907812_29907963 | 0.25 |
ENSG00000199187 |
. |
5457 |
0.13 |
| chr5_14038564_14038715 | 0.25 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
93987 |
0.09 |
| chr1_10961180_10961385 | 0.25 |
C1orf127 |
chromosome 1 open reading frame 127 |
46645 |
0.12 |
| chr5_65107256_65107407 | 0.25 |
NLN |
neurolysin (metallopeptidase M3 family) |
23141 |
0.22 |
| chr1_10891158_10891309 | 0.25 |
CASZ1 |
castor zinc finger 1 |
34526 |
0.18 |
| chr9_127457900_127458051 | 0.24 |
ENSG00000207737 |
. |
1986 |
0.26 |
| chr1_59491106_59491398 | 0.24 |
JUN |
jun proto-oncogene |
241467 |
0.02 |
| chr6_10633412_10633876 | 0.24 |
GCNT6 |
glucosaminyl (N-acetyl) transferase 6 |
349 |
0.86 |
| chr8_101774241_101774392 | 0.24 |
ENSG00000202001 |
. |
6893 |
0.17 |
| chr1_202557788_202557939 | 0.23 |
RP11-569A11.1 |
|
15533 |
0.2 |
| chr15_79621555_79621707 | 0.23 |
ENSG00000266543 |
. |
9441 |
0.2 |
| chr21_43084452_43084603 | 0.23 |
LINC00112 |
long intergenic non-protein coding RNA 112 |
52069 |
0.12 |
| chr22_37669382_37669767 | 0.23 |
CYTH4 |
cytohesin 4 |
8494 |
0.16 |
| chr11_65259042_65259542 | 0.23 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
33256 |
0.08 |
| chr7_137457647_137457798 | 0.23 |
DGKI |
diacylglycerol kinase, iota |
73560 |
0.12 |
| chr6_100731277_100731428 | 0.23 |
RP1-121G13.2 |
|
143642 |
0.05 |
| chr6_43090954_43091105 | 0.23 |
PTK7 |
protein tyrosine kinase 7 |
7300 |
0.13 |
| chr22_27288381_27288532 | 0.23 |
ENSG00000200443 |
. |
143394 |
0.05 |
| chr5_149401334_149401630 | 0.23 |
ENSG00000238369 |
. |
7533 |
0.14 |
| chr13_109567579_109568598 | 0.23 |
MYO16 |
myosin XVI |
29571 |
0.26 |
| chr1_95163966_95164117 | 0.23 |
ENSG00000263526 |
. |
47415 |
0.17 |
| chr1_214731522_214731884 | 0.22 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
7137 |
0.28 |
| chr8_143757384_143757535 | 0.22 |
PSCA |
prostate stem cell antigen |
4415 |
0.13 |
| chr9_132007671_132007822 | 0.22 |
ENSG00000220992 |
. |
14750 |
0.17 |
| chr22_47330919_47331070 | 0.22 |
ENSG00000221672 |
. |
87191 |
0.09 |
| chr18_46463843_46464371 | 0.22 |
SMAD7 |
SMAD family member 7 |
10768 |
0.25 |
| chr2_153271980_153272131 | 0.22 |
FMNL2 |
formin-like 2 |
80304 |
0.11 |
| chr15_40577810_40578209 | 0.22 |
ANKRD63 |
ankyrin repeat domain 63 |
3222 |
0.12 |
| chr1_154701622_154701930 | 0.22 |
ADAR |
adenosine deaminase, RNA-specific |
101302 |
0.05 |
| chr4_148698718_148698869 | 0.22 |
ENSG00000264274 |
. |
4953 |
0.21 |
| chr3_48603652_48603803 | 0.22 |
UCN2 |
urocortin 2 |
2521 |
0.14 |
| chr5_52105602_52106304 | 0.21 |
CTD-2288O8.1 |
|
22093 |
0.18 |
| chr7_24826656_24826935 | 0.21 |
DFNA5 |
deafness, autosomal dominant 5 |
17551 |
0.26 |
| chr17_2786695_2787071 | 0.21 |
CTD-3060P21.1 |
|
82306 |
0.07 |
| chr2_238410967_238411392 | 0.21 |
MLPH |
melanophilin |
15260 |
0.18 |
| chr11_9010825_9011174 | 0.21 |
NRIP3 |
nuclear receptor interacting protein 3 |
3696 |
0.14 |
| chr2_60743198_60743767 | 0.21 |
AC009970.1 |
|
20661 |
0.21 |
| chr1_27325551_27325702 | 0.21 |
TRNP1 |
TMF1-regulated nuclear protein 1 |
4816 |
0.17 |
| chr17_28111866_28112121 | 0.21 |
ENSG00000252657 |
. |
7356 |
0.17 |
| chr5_153003729_153003880 | 0.21 |
ENSG00000242976 |
. |
131138 |
0.05 |
| chr19_49071957_49072108 | 0.21 |
SULT2B1 |
sulfotransferase family, cytosolic, 2B, member 1 |
6765 |
0.11 |
| chr21_33836630_33836895 | 0.21 |
EVA1C |
eva-1 homolog C (C. elegans) |
36120 |
0.13 |
| chr21_38936455_38936763 | 0.21 |
AP001421.1 |
Uncharacterized protein |
47869 |
0.16 |
| chr21_38711029_38711180 | 0.21 |
AP001437.1 |
|
27130 |
0.16 |
| chr6_107009204_107009507 | 0.21 |
AIM1 |
absent in melanoma 1 |
20326 |
0.18 |
| chr16_57841238_57841389 | 0.21 |
CTD-2600O9.1 |
uncharacterized protein LOC388282 |
3236 |
0.18 |
| chr20_3824293_3824586 | 0.21 |
MAVS |
mitochondrial antiviral signaling protein |
3048 |
0.18 |
| chr2_169429731_169430128 | 0.21 |
ENSG00000265694 |
. |
9524 |
0.23 |
| chr5_176040795_176040946 | 0.21 |
GPRIN1 |
G protein regulated inducer of neurite outgrowth 1 |
3736 |
0.14 |
| chr5_106767835_106767986 | 0.21 |
EFNA5 |
ephrin-A5 |
238418 |
0.02 |
| chr5_159609192_159609343 | 0.21 |
FABP6 |
fatty acid binding protein 6, ileal |
5107 |
0.2 |
| chr6_158902774_158902925 | 0.20 |
ENSG00000243373 |
. |
44020 |
0.14 |
| chr11_130903385_130903536 | 0.20 |
ENSG00000242673 |
. |
29136 |
0.23 |
| chr14_75355002_75355210 | 0.20 |
DLST |
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
6447 |
0.15 |
| chr5_123932447_123932800 | 0.20 |
RP11-436H11.2 |
|
131901 |
0.05 |
| chr20_52407741_52407914 | 0.20 |
ENSG00000238468 |
. |
122530 |
0.05 |
| chr16_10678700_10678851 | 0.20 |
EMP2 |
epithelial membrane protein 2 |
4220 |
0.21 |
| chr5_14268148_14268359 | 0.20 |
TRIO |
trio Rho guanine nucleotide exchange factor |
22833 |
0.28 |
| chr19_13163011_13163312 | 0.20 |
AC007787.2 |
|
19027 |
0.09 |
| chr15_36720781_36720932 | 0.20 |
C15orf41 |
chromosome 15 open reading frame 41 |
150956 |
0.05 |
| chr3_11642284_11642435 | 0.20 |
VGLL4 |
vestigial like 4 (Drosophila) |
3696 |
0.24 |
| chr12_132356722_132356873 | 0.20 |
ULK1 |
unc-51 like autophagy activating kinase 1 |
22399 |
0.14 |
| chr15_71881190_71881341 | 0.20 |
THSD4 |
thrombospondin, type I, domain containing 4 |
41690 |
0.19 |
| chr16_11295567_11296301 | 0.20 |
RMI2 |
RecQ mediated genome instability 2 |
47572 |
0.08 |
| chr3_71651118_71651323 | 0.20 |
FOXP1 |
forkhead box P1 |
18080 |
0.2 |
| chr5_135347184_135347335 | 0.20 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
17325 |
0.2 |
| chr6_56526637_56526788 | 0.20 |
DST |
dystonin |
18918 |
0.28 |
| chr12_24893513_24893767 | 0.19 |
ENSG00000240481 |
. |
33918 |
0.21 |
| chr12_124997368_124997619 | 0.19 |
NCOR2 |
nuclear receptor corepressor 2 |
5347 |
0.33 |
| chr15_99664062_99664434 | 0.19 |
RP11-6O2.4 |
|
7763 |
0.14 |
| chr10_29273394_29273655 | 0.19 |
ENSG00000199402 |
. |
110088 |
0.07 |
| chr14_103603313_103603536 | 0.19 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
4339 |
0.2 |
| chr11_104005552_104005703 | 0.19 |
PDGFD |
platelet derived growth factor D |
29208 |
0.23 |
| chr7_55260410_55260561 | 0.19 |
EGFR-AS1 |
EGFR antisense RNA 1 |
3858 |
0.34 |
| chr16_14404359_14404586 | 0.19 |
ENSG00000199130 |
. |
1330 |
0.44 |
| chr5_72969116_72969415 | 0.19 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
11392 |
0.24 |
| chr10_70982539_70983066 | 0.19 |
HKDC1 |
hexokinase domain containing 1 |
2743 |
0.22 |
| chr2_110859211_110859419 | 0.19 |
MALL |
mal, T-cell differentiation protein-like |
14284 |
0.15 |
| chr9_84174156_84174449 | 0.19 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
54135 |
0.17 |
| chr5_148513977_148514128 | 0.19 |
ABLIM3 |
actin binding LIM protein family, member 3 |
6994 |
0.18 |
| chr17_47533894_47534339 | 0.19 |
NGFR |
nerve growth factor receptor |
38539 |
0.1 |
| chr18_21978734_21978885 | 0.19 |
OSBPL1A |
oxysterol binding protein-like 1A |
986 |
0.48 |
| chr1_155223556_155224415 | 0.19 |
FAM189B |
family with sequence similarity 189, member B |
714 |
0.38 |
| chr18_10035524_10035675 | 0.19 |
ENSG00000263630 |
. |
30405 |
0.21 |
| chr15_67467838_67468111 | 0.19 |
SMAD3 |
SMAD family member 3 |
9105 |
0.22 |
| chr19_4794956_4795151 | 0.19 |
AC005523.2 |
|
514 |
0.66 |
| chr16_88298714_88299235 | 0.19 |
ZNF469 |
zinc finger protein 469 |
194905 |
0.02 |
| chr20_34673805_34674098 | 0.19 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
5475 |
0.21 |
| chr9_127023759_127023910 | 0.19 |
NEK6 |
NIMA-related kinase 6 |
82 |
0.83 |
| chr7_91476158_91476309 | 0.19 |
MTERF |
mitochondrial transcription termination factor |
33744 |
0.22 |
| chr19_16174815_16174966 | 0.19 |
TPM4 |
tropomyosin 4 |
2941 |
0.24 |
| chr18_59658543_59658754 | 0.18 |
RNF152 |
ring finger protein 152 |
97184 |
0.08 |
| chr12_68958912_68959888 | 0.18 |
RAP1B |
RAP1B, member of RAS oncogene family |
45219 |
0.16 |
| chr2_29850371_29850522 | 0.18 |
ENSG00000242699 |
. |
53743 |
0.16 |
| chr22_41961672_41961960 | 0.18 |
CSDC2 |
cold shock domain containing C2, RNA binding |
5049 |
0.14 |
| chr1_200746157_200746308 | 0.18 |
CAMSAP2 |
calmodulin regulated spectrin-associated protein family, member 2 |
37325 |
0.18 |
| chr18_55758646_55758797 | 0.18 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
44263 |
0.17 |
| chr9_113515310_113515461 | 0.18 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
22242 |
0.25 |
| chr5_141742469_141742735 | 0.18 |
AC005592.2 |
|
9494 |
0.25 |
| chr7_155088256_155088792 | 0.18 |
INSIG1 |
insulin induced gene 1 |
962 |
0.62 |
| chr15_52369332_52369483 | 0.18 |
CTD-2184D3.5 |
|
23313 |
0.13 |
| chr22_47226754_47226905 | 0.18 |
ENSG00000221672 |
. |
16974 |
0.22 |
| chr12_112741045_112741373 | 0.18 |
ENSG00000201428 |
. |
36328 |
0.15 |
| chr11_93470751_93471427 | 0.18 |
ENSG00000210825 |
. |
2687 |
0.08 |
| chr2_43152306_43152732 | 0.18 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
132787 |
0.05 |
| chr4_148752956_148753135 | 0.18 |
ENSG00000240014 |
. |
22968 |
0.19 |
| chr19_18681741_18681892 | 0.18 |
UBA52 |
ubiquitin A-52 residue ribosomal protein fusion product 1 |
724 |
0.4 |
| chr19_49071674_49071825 | 0.18 |
SULT2B1 |
sulfotransferase family, cytosolic, 2B, member 1 |
7048 |
0.11 |
| chr1_114665539_114665690 | 0.18 |
SYT6 |
synaptotagmin VI |
29448 |
0.22 |
| chr19_38794620_38794771 | 0.18 |
C19orf33 |
chromosome 19 open reading frame 33 |
106 |
0.92 |
| chr17_42622509_42622660 | 0.18 |
FZD2 |
frizzled family receptor 2 |
12341 |
0.18 |
| chr9_16204302_16204453 | 0.18 |
C9orf92 |
chromosome 9 open reading frame 92 |
11520 |
0.31 |
| chr9_135506787_135506938 | 0.18 |
DDX31 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
32025 |
0.15 |
| chr10_6261359_6261510 | 0.18 |
PFKFB3 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
1933 |
0.3 |
| chr1_26610395_26611734 | 0.18 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
4451 |
0.13 |
| chr6_130023402_130023553 | 0.18 |
ARHGAP18 |
Rho GTPase activating protein 18 |
7893 |
0.27 |
| chr2_39869646_39869797 | 0.18 |
TMEM178A |
transmembrane protein 178A |
23338 |
0.24 |
| chr9_97682173_97682324 | 0.18 |
RP11-54O15.3 |
|
14980 |
0.17 |
| chr2_25570321_25570568 | 0.17 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
4985 |
0.24 |
| chr18_74452398_74452589 | 0.17 |
ENSG00000252097 |
. |
60237 |
0.13 |
| chr18_8122134_8122285 | 0.17 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
8640 |
0.24 |
| chr19_576051_576313 | 0.17 |
BSG |
basigin (Ok blood group) |
961 |
0.34 |
| chr2_235586595_235586746 | 0.17 |
ARL4C |
ADP-ribosylation factor-like 4C |
180973 |
0.03 |
| chr16_9141478_9141964 | 0.17 |
C16orf72 |
chromosome 16 open reading frame 72 |
43784 |
0.12 |
| chr22_46402793_46403252 | 0.17 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
30013 |
0.1 |
| chr20_25290765_25291165 | 0.17 |
ABHD12 |
abhydrolase domain containing 12 |
831 |
0.64 |
| chr5_10555649_10555800 | 0.17 |
ANKRD33B |
ankyrin repeat domain 33B |
8856 |
0.2 |
| chr8_37373070_37373278 | 0.17 |
RP11-150O12.6 |
|
1365 |
0.56 |
| chr8_71125713_71126274 | 0.17 |
NCOA2 |
nuclear receptor coactivator 2 |
31617 |
0.21 |
| chr15_59460757_59460908 | 0.17 |
ENSG00000253030 |
. |
2629 |
0.17 |
| chr5_109201537_109201688 | 0.17 |
AC011366.3 |
Uncharacterized protein |
17271 |
0.29 |
| chr18_9743354_9743731 | 0.17 |
RAB31 |
RAB31, member RAS oncogene family |
35380 |
0.17 |
| chr2_20696028_20696179 | 0.17 |
ENSG00000200829 |
. |
15498 |
0.21 |
| chr4_53791904_53792055 | 0.17 |
RP11-752D24.2 |
|
19837 |
0.23 |
| chr4_48175111_48175594 | 0.17 |
ENSG00000202014 |
. |
19901 |
0.18 |
| chr11_69069206_69069429 | 0.17 |
MYEOV |
myeloma overexpressed |
7692 |
0.29 |
| chr17_78801730_78801881 | 0.17 |
RP11-28G8.1 |
|
22373 |
0.2 |
| chr1_17525565_17525716 | 0.17 |
PADI1 |
peptidyl arginine deiminase, type I |
5981 |
0.19 |
| chr2_206599303_206599570 | 0.17 |
AC007362.3 |
|
29241 |
0.21 |
| chr16_4994791_4994942 | 0.17 |
ENSG00000200980 |
. |
7026 |
0.15 |
| chr1_41568152_41568303 | 0.17 |
SCMH1 |
sex comb on midleg homolog 1 (Drosophila) |
57378 |
0.13 |
| chr5_14165682_14165833 | 0.17 |
TRIO |
trio Rho guanine nucleotide exchange factor |
18150 |
0.3 |
| chr2_20696298_20696449 | 0.17 |
ENSG00000200829 |
. |
15768 |
0.21 |
| chr22_44756801_44756952 | 0.17 |
RP1-32I10.10 |
Uncharacterized protein |
4555 |
0.29 |
| chr17_60780537_60780688 | 0.17 |
RP11-156L14.1 |
|
2955 |
0.22 |
| chr12_52074390_52074541 | 0.17 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
5742 |
0.28 |
| chr1_222060065_222060216 | 0.17 |
ENSG00000200033 |
. |
49361 |
0.18 |
| chr2_47420449_47420733 | 0.17 |
CALM2 |
calmodulin 2 (phosphorylase kinase, delta) |
16851 |
0.2 |
| chr8_22779071_22779222 | 0.17 |
PEBP4 |
phosphatidylethanolamine-binding protein 4 |
6275 |
0.16 |
| chr12_117039449_117039758 | 0.17 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
25947 |
0.23 |
| chr20_19601854_19602005 | 0.16 |
ENSG00000216017 |
. |
31098 |
0.2 |
| chr16_4368044_4368195 | 0.16 |
GLIS2 |
GLIS family zinc finger 2 |
3357 |
0.17 |
| chr7_35635806_35635957 | 0.16 |
HERPUD2 |
HERPUD family member 2 |
98295 |
0.08 |
| chr4_102077826_102078189 | 0.16 |
ENSG00000221265 |
. |
173564 |
0.03 |
| chr5_14268406_14268609 | 0.16 |
TRIO |
trio Rho guanine nucleotide exchange factor |
22579 |
0.28 |
| chr12_77425511_77425662 | 0.16 |
E2F7 |
E2F transcription factor 7 |
18940 |
0.27 |
| chr3_196001577_196001860 | 0.16 |
PCYT1A |
phosphate cytidylyltransferase 1, choline, alpha |
4306 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.2 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.2 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0090594 | wound healing involved in inflammatory response(GO:0002246) inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.0 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.1 | GO:0031958 | corticosteroid receptor signaling pathway(GO:0031958) glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.1 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.0 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.0 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.2 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0060087 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.0 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0047115 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |