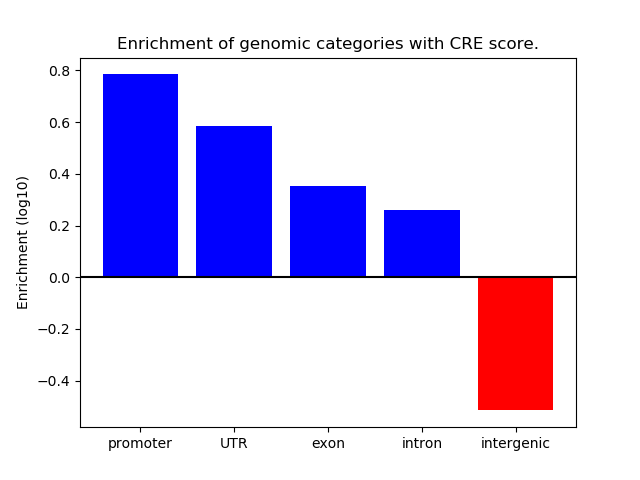
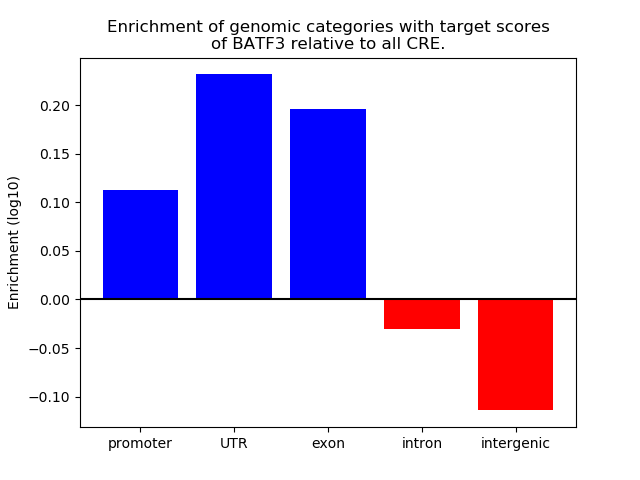
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
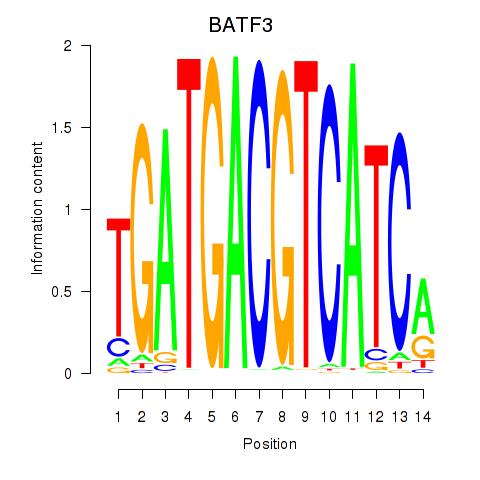
Results for BATF3
Z-value: 0.27
Transcription factors associated with BATF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BATF3
|
ENSG00000123685.4 | basic leucine zipper ATF-like transcription factor 3 |
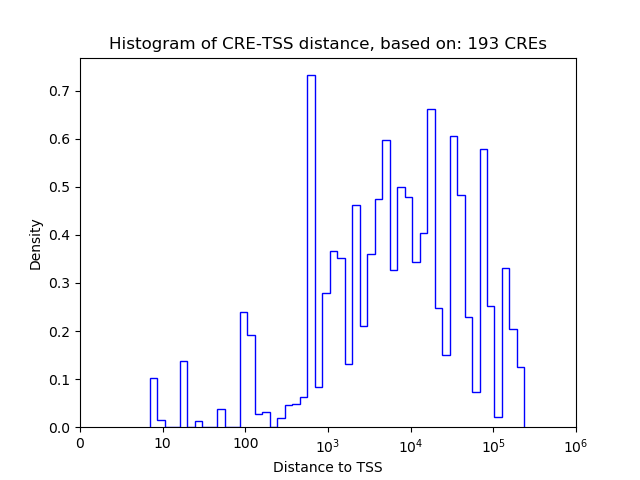
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_212911456_212911607 | BATF3 | 38204 | 0.119129 | 0.82 | 7.1e-03 | Click! |
| chr1_212884026_212884300 | BATF3 | 10836 | 0.168773 | 0.55 | 1.3e-01 | Click! |
| chr1_212894826_212895572 | BATF3 | 21872 | 0.151734 | -0.50 | 1.7e-01 | Click! |
| chr1_212874351_212874643 | BATF3 | 1170 | 0.452951 | -0.43 | 2.5e-01 | Click! |
| chr1_212901261_212901450 | BATF3 | 28028 | 0.140262 | 0.38 | 3.1e-01 | Click! |
Activity of the BATF3 motif across conditions
Conditions sorted by the z-value of the BATF3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
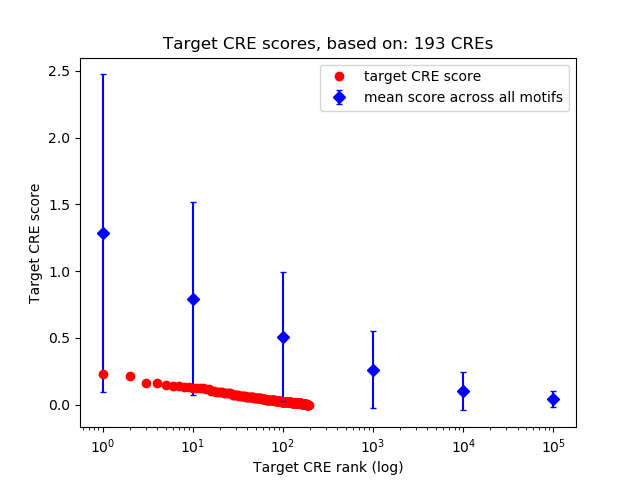
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_156696862_156697210 | 0.23 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
674 |
0.62 |
| chr2_144280347_144280652 | 0.22 |
AC096558.1 |
|
42141 |
0.16 |
| chr4_170581231_170581398 | 0.16 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
101 |
0.98 |
| chr9_6915729_6915880 | 0.16 |
KDM4C |
lysine (K)-specific demethylase 4C |
9697 |
0.24 |
| chr5_177900691_177901194 | 0.15 |
CTB-26E19.1 |
|
30847 |
0.19 |
| chr21_18983750_18984027 | 0.14 |
BTG3 |
BTG family, member 3 |
1274 |
0.5 |
| chr17_1305657_1305808 | 0.14 |
YWHAE |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon |
2060 |
0.29 |
| chr6_158306516_158306953 | 0.13 |
RP3-403L10.3 |
|
7084 |
0.18 |
| chr13_30747324_30747475 | 0.13 |
ENSG00000266816 |
. |
33437 |
0.22 |
| chr4_185069136_185069287 | 0.13 |
ENPP6 |
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
69903 |
0.11 |
| chr3_45911190_45911341 | 0.13 |
CCR9 |
chemokine (C-C motif) receptor 9 |
16731 |
0.15 |
| chr1_186798987_186799161 | 0.13 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
952 |
0.72 |
| chrX_129318276_129318427 | 0.12 |
RAB33A |
RAB33A, member RAS oncogene family |
12728 |
0.19 |
| chr3_16843338_16843591 | 0.12 |
PLCL2 |
phospholipase C-like 2 |
82988 |
0.1 |
| chr3_46935271_46935422 | 0.12 |
AC109583.1 |
Uncharacterized protein |
5175 |
0.16 |
| chr6_135518184_135518665 | 0.10 |
MYB-AS1 |
MYB antisense RNA 1 |
1291 |
0.44 |
| chr10_8284374_8284525 | 0.10 |
GATA3 |
GATA binding protein 3 |
187680 |
0.03 |
| chr9_100395113_100395272 | 0.10 |
TSTD2 |
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 |
665 |
0.42 |
| chr3_134092582_134092733 | 0.09 |
AMOTL2 |
angiomotin like 2 |
18 |
0.98 |
| chr11_59279292_59279443 | 0.09 |
OR4D9 |
olfactory receptor, family 4, subfamily D, member 9 |
3019 |
0.16 |
| chr7_100464718_100465052 | 0.09 |
TRIP6 |
thyroid hormone receptor interactor 6 |
125 |
0.92 |
| chr14_100112045_100112211 | 0.09 |
HHIPL1 |
HHIP-like 1 |
648 |
0.71 |
| chrX_19770106_19770364 | 0.09 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
4414 |
0.34 |
| chr3_32451109_32451284 | 0.09 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
17665 |
0.23 |
| chr2_111740835_111740986 | 0.09 |
ACOXL |
acyl-CoA oxidase-like |
3865 |
0.35 |
| chr5_64331605_64331756 | 0.08 |
ENSG00000207439 |
. |
87516 |
0.1 |
| chr17_62032388_62032539 | 0.08 |
SCN4A |
sodium channel, voltage-gated, type IV, alpha subunit |
17815 |
0.11 |
| chr1_198904060_198904298 | 0.08 |
ENSG00000207759 |
. |
75897 |
0.11 |
| chr20_25007310_25007600 | 0.07 |
ACSS1 |
acyl-CoA synthetase short-chain family member 1 |
5853 |
0.21 |
| chr13_111227788_111227989 | 0.07 |
RAB20 |
RAB20, member RAS oncogene family |
13808 |
0.2 |
| chr10_82181789_82181952 | 0.07 |
FAM213A |
family with sequence similarity 213, member A |
2116 |
0.32 |
| chr3_62304486_62304637 | 0.07 |
ENSG00000241472 |
. |
6 |
0.57 |
| chr2_219926016_219926467 | 0.06 |
IHH |
indian hedgehog |
1052 |
0.34 |
| chr8_6114669_6114820 | 0.06 |
RP11-115C21.2 |
|
149319 |
0.04 |
| chr13_33646591_33646742 | 0.06 |
ENSG00000221677 |
. |
31177 |
0.19 |
| chr6_156968465_156968883 | 0.06 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
130389 |
0.05 |
| chr11_45831767_45831918 | 0.06 |
SLC35C1 |
solute carrier family 35 (GDP-fucose transporter), member C1 |
5142 |
0.18 |
| chr1_94366888_94367039 | 0.06 |
GCLM |
glutamate-cysteine ligase, modifier subunit |
8003 |
0.19 |
| chr10_133747286_133747437 | 0.06 |
PPP2R2D |
protein phosphatase 2, regulatory subunit B, delta |
594 |
0.81 |
| chr19_50861782_50862295 | 0.06 |
NAPSA |
napsin A aspartic peptidase |
6844 |
0.09 |
| chr21_46001951_46002102 | 0.06 |
KRTAP10-5 |
keratin associated protein 10-5 |
1545 |
0.17 |
| chr15_98224143_98224294 | 0.06 |
LINC00923 |
long intergenic non-protein coding RNA 923 |
193562 |
0.03 |
| chr4_77212822_77212973 | 0.06 |
FAM47E-STBD1 |
FAM47E-STBD1 readthrough |
14282 |
0.17 |
| chr15_81582433_81582671 | 0.06 |
IL16 |
interleukin 16 |
6702 |
0.22 |
| chr8_82100250_82100582 | 0.06 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
76113 |
0.1 |
| chr11_60880202_60880378 | 0.06 |
CD5 |
CD5 molecule |
10309 |
0.18 |
| chr14_92346455_92346606 | 0.05 |
FBLN5 |
fibulin 5 |
2144 |
0.33 |
| chr20_46369136_46369287 | 0.05 |
SULF2 |
sulfatase 2 |
45022 |
0.15 |
| chr2_68612447_68612608 | 0.05 |
AC015969.3 |
|
19811 |
0.16 |
| chr6_82569584_82569847 | 0.05 |
ENSG00000206886 |
. |
95974 |
0.08 |
| chr19_15579754_15579917 | 0.05 |
RASAL3 |
RAS protein activator like 3 |
4453 |
0.14 |
| chr12_56731704_56731855 | 0.05 |
IL23A |
interleukin 23, alpha subunit p19 |
884 |
0.33 |
| chr19_2775823_2776196 | 0.05 |
SGTA |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
5230 |
0.12 |
| chr5_176569109_176569306 | 0.05 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
7102 |
0.21 |
| chr5_75748396_75748547 | 0.05 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
48222 |
0.17 |
| chr21_34707233_34707384 | 0.05 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
10030 |
0.18 |
| chr21_46898650_46899022 | 0.05 |
COL18A1 |
collagen, type XVIII, alpha 1 |
11353 |
0.2 |
| chr5_59437071_59437222 | 0.04 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
44279 |
0.19 |
| chr6_16569798_16569949 | 0.04 |
ENSG00000265642 |
. |
141119 |
0.05 |
| chrX_109249032_109249228 | 0.04 |
TMEM164 |
transmembrane protein 164 |
2787 |
0.34 |
| chr6_16749772_16749987 | 0.04 |
RP1-151F17.1 |
|
11490 |
0.24 |
| chr12_3857937_3858128 | 0.04 |
EFCAB4B |
EF-hand calcium binding domain 4B |
4231 |
0.27 |
| chr1_32810829_32811029 | 0.04 |
TSSK3 |
testis-specific serine kinase 3 |
8126 |
0.12 |
| chr10_48289621_48289908 | 0.04 |
ANXA8 |
annexin A8 |
34411 |
0.11 |
| chrX_135724373_135724536 | 0.04 |
ENSG00000233093 |
. |
2628 |
0.24 |
| chr2_51452608_51452759 | 0.04 |
NRXN1 |
neurexin 1 |
193009 |
0.03 |
| chr17_79328867_79329074 | 0.04 |
TMEM105 |
transmembrane protein 105 |
24496 |
0.11 |
| chr6_73904661_73904812 | 0.04 |
KHDC1P1 |
KH homology domain containing 1 pseudogene 1 |
15067 |
0.15 |
| chr17_65505786_65506259 | 0.04 |
CTD-2653B5.1 |
|
14575 |
0.17 |
| chr20_56967400_56967551 | 0.04 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
3106 |
0.31 |
| chr10_103608236_103608387 | 0.03 |
KCNIP2 |
Kv channel interacting protein 2 |
4634 |
0.19 |
| chr9_92110830_92111100 | 0.03 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
1382 |
0.41 |
| chr5_61612133_61612284 | 0.03 |
KIF2A |
kinesin heavy chain member 2A |
4568 |
0.3 |
| chr22_30662165_30662316 | 0.03 |
OSM |
oncostatin M |
433 |
0.7 |
| chr3_101555850_101556001 | 0.03 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
9088 |
0.19 |
| chr2_217641332_217641483 | 0.03 |
ENSG00000222832 |
. |
16916 |
0.18 |
| chr2_198575264_198575450 | 0.03 |
MARS2 |
methionyl-tRNA synthetase 2, mitochondrial |
5270 |
0.2 |
| chr14_58605715_58605866 | 0.03 |
C14orf37 |
chromosome 14 open reading frame 37 |
13141 |
0.19 |
| chr13_52209380_52209601 | 0.03 |
ENSG00000242893 |
. |
41139 |
0.12 |
| chr17_75531537_75531688 | 0.03 |
SEPT9 |
septin 9 |
53617 |
0.12 |
| chr11_77785895_77786046 | 0.03 |
NDUFC2 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
4895 |
0.12 |
| chr6_36076165_36076316 | 0.03 |
MAPK13 |
mitogen-activated protein kinase 13 |
21858 |
0.16 |
| chr22_39745336_39745494 | 0.03 |
SYNGR1 |
synaptogyrin 1 |
515 |
0.69 |
| chr3_193461847_193462134 | 0.03 |
ENSG00000243991 |
. |
3480 |
0.32 |
| chr16_50742452_50742934 | 0.03 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
3412 |
0.14 |
| chr11_12695755_12695947 | 0.03 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
118 |
0.98 |
| chr11_77524472_77524623 | 0.03 |
RSF1 |
remodeling and spacing factor 1 |
7205 |
0.15 |
| chr8_30762092_30762243 | 0.03 |
TEX15 |
testis expressed 15 |
55559 |
0.14 |
| chr17_76749954_76750222 | 0.03 |
CYTH1 |
cytohesin 1 |
17116 |
0.18 |
| chr8_27222091_27222616 | 0.03 |
PTK2B |
protein tyrosine kinase 2 beta |
15815 |
0.21 |
| chr7_138393128_138393279 | 0.03 |
SVOPL |
SVOP-like |
7106 |
0.24 |
| chr15_48469683_48470033 | 0.03 |
MYEF2 |
myelin expression factor 2 |
692 |
0.6 |
| chr9_124091436_124091643 | 0.03 |
GSN |
gelsolin |
2679 |
0.21 |
| chrX_56791597_56791859 | 0.03 |
ENSG00000204272 |
. |
36036 |
0.23 |
| chr13_97876107_97876342 | 0.02 |
MBNL2 |
muscleblind-like splicing regulator 2 |
1615 |
0.53 |
| chr7_132350846_132351136 | 0.02 |
AC009365.3 |
|
17438 |
0.2 |
| chr20_61808362_61808898 | 0.02 |
ENSG00000207598 |
. |
1222 |
0.4 |
| chr8_74792715_74792866 | 0.02 |
UBE2W |
ubiquitin-conjugating enzyme E2W (putative) |
1645 |
0.41 |
| chr10_73520995_73521418 | 0.02 |
C10orf54 |
chromosome 10 open reading frame 54 |
3819 |
0.23 |
| chr5_87963475_87963729 | 0.02 |
ENSG00000245526 |
. |
845 |
0.63 |
| chr1_40860718_40861074 | 0.02 |
SMAP2 |
small ArfGAP2 |
1611 |
0.36 |
| chr17_62141355_62141640 | 0.02 |
ICAM2 |
intercellular adhesion molecule 2 |
43503 |
0.11 |
| chr13_113664073_113664237 | 0.02 |
RP11-120K24.5 |
|
170 |
0.9 |
| chr13_41139475_41139713 | 0.02 |
AL133318.1 |
Uncharacterized protein |
28271 |
0.22 |
| chr10_50452094_50452371 | 0.02 |
C10orf71-AS1 |
C10orf71 antisense RNA 1 |
54593 |
0.1 |
| chr7_8297929_8298080 | 0.02 |
ICA1 |
islet cell autoantigen 1, 69kDa |
3678 |
0.21 |
| chr3_38079216_38079504 | 0.02 |
DLEC1 |
deleted in lung and esophageal cancer 1 |
1336 |
0.35 |
| chr5_158176936_158177087 | 0.02 |
CTD-2363C16.1 |
|
233003 |
0.02 |
| chr21_44184193_44184344 | 0.02 |
AP001627.1 |
|
22400 |
0.2 |
| chr6_2111445_2111677 | 0.02 |
GMDS |
GDP-mannose 4,6-dehydratase |
64664 |
0.15 |
| chr13_107862893_107863044 | 0.02 |
ENSG00000221316 |
. |
89587 |
0.09 |
| chr4_2284128_2284303 | 0.02 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
5227 |
0.14 |
| chr1_205779229_205779380 | 0.02 |
SLC41A1 |
solute carrier family 41 (magnesium transporter), member 1 |
3000 |
0.23 |
| chrX_135848042_135848429 | 0.02 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
1267 |
0.45 |
| chr17_62972703_62972949 | 0.02 |
AMZ2P1 |
archaelysin family metallopeptidase 2 pseudogene 1 |
3191 |
0.2 |
| chr19_14117646_14117797 | 0.02 |
RFX1 |
regulatory factor X, 1 (influences HLA class II expression) |
624 |
0.56 |
| chr7_74837064_74837215 | 0.02 |
AC004878.2 |
|
18173 |
0.13 |
| chr17_63048049_63048280 | 0.02 |
RP11-583F2.5 |
|
137 |
0.96 |
| chr10_126672174_126672466 | 0.02 |
CTBP2 |
C-terminal binding protein 2 |
22262 |
0.16 |
| chr18_3277346_3277526 | 0.02 |
MYL12B |
myosin, light chain 12B, regulatory |
14482 |
0.16 |
| chr12_133022629_133022780 | 0.02 |
MUC8 |
mucin 8 |
28022 |
0.17 |
| chr9_245521_245672 | 0.02 |
RP11-59O6.3 |
|
27406 |
0.15 |
| chr1_9790555_9790706 | 0.02 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
12503 |
0.17 |
| chr8_10697366_10697517 | 0.02 |
PINX1 |
PIN2/TERF1 interacting, telomerase inhibitor 1 |
55 |
0.51 |
| chr15_65576232_65576450 | 0.02 |
ENSG00000206903 |
. |
1588 |
0.26 |
| chr14_76010271_76010422 | 0.02 |
BATF |
basic leucine zipper transcription factor, ATF-like |
21443 |
0.15 |
| chr19_5687992_5688143 | 0.02 |
RPL36 |
ribosomal protein L36 |
2140 |
0.17 |
| chr22_24555252_24555403 | 0.02 |
CABIN1 |
calcineurin binding protein 1 |
3521 |
0.21 |
| chr2_144075020_144075171 | 0.02 |
RP11-190J23.1 |
|
145354 |
0.04 |
| chr7_38309731_38309882 | 0.02 |
STARD3NL |
STARD3 N-terminal like |
91809 |
0.09 |
| chr2_228312998_228313149 | 0.01 |
ENSG00000266382 |
. |
23775 |
0.17 |
| chr21_27457592_27457743 | 0.01 |
APP |
amyloid beta (A4) precursor protein |
4710 |
0.29 |
| chr14_22457517_22457668 | 0.01 |
ENSG00000238634 |
. |
153295 |
0.04 |
| chr19_45625479_45625630 | 0.01 |
AC005757.6 |
|
14681 |
0.09 |
| chr22_45664092_45664243 | 0.01 |
UPK3A |
uroplakin 3A |
16696 |
0.16 |
| chr7_76266405_76266556 | 0.01 |
POMZP3 |
POM121 and ZP3 fusion |
9922 |
0.22 |
| chr17_2516877_2517028 | 0.01 |
PAFAH1B1 |
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) |
19912 |
0.14 |
| chr1_244396134_244396285 | 0.01 |
C1orf100 |
chromosome 1 open reading frame 100 |
119728 |
0.05 |
| chr20_62580456_62580607 | 0.01 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
1948 |
0.13 |
| chr7_75081723_75081874 | 0.01 |
POM121C |
POM121 transmembrane nucleoporin C |
9805 |
0.13 |
| chr1_167193026_167193202 | 0.01 |
POU2F1 |
POU class 2 homeobox 1 |
2971 |
0.23 |
| chr11_66059435_66059843 | 0.01 |
TMEM151A |
transmembrane protein 151A |
298 |
0.71 |
| chr1_22380057_22380208 | 0.01 |
CDC42 |
cell division cycle 42 |
342 |
0.82 |
| chr6_32164359_32164510 | 0.01 |
NOTCH4 |
notch 4 |
921 |
0.28 |
| chr9_80362813_80362964 | 0.01 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
75027 |
0.11 |
| chr11_12071055_12071206 | 0.01 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
39814 |
0.17 |
| chr16_90021030_90021181 | 0.01 |
DEF8 |
differentially expressed in FDCP 8 homolog (mouse) |
525 |
0.64 |
| chr7_50535965_50536168 | 0.01 |
FIGNL1 |
fidgetin-like 1 |
17978 |
0.2 |
| chr3_140693787_140693938 | 0.01 |
RP11-231L11.3 |
|
2276 |
0.36 |
| chr22_32805402_32805689 | 0.01 |
RTCB |
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
2679 |
0.22 |
| chr3_107646211_107646473 | 0.01 |
BBX |
bobby sox homolog (Drosophila) |
128904 |
0.06 |
| chr17_5365759_5365910 | 0.01 |
DHX33 |
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
6234 |
0.12 |
| chr19_11074648_11074881 | 0.01 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
2767 |
0.19 |
| chr17_75425276_75425533 | 0.01 |
SEPT9 |
septin 9 |
2366 |
0.24 |
| chr1_47060894_47061171 | 0.01 |
MKNK1 |
MAP kinase interacting serine/threonine kinase 1 |
301 |
0.87 |
| chr6_37891842_37892090 | 0.01 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
5769 |
0.23 |
| chr5_139930646_139930797 | 0.01 |
SRA1 |
steroid receptor RNA activator 1 |
8 |
0.94 |
| chr2_37062565_37062716 | 0.01 |
AC007382.1 |
Uncharacterized protein |
5890 |
0.23 |
| chr14_25103522_25103673 | 0.01 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
124 |
0.96 |
| chr10_112016537_112016688 | 0.01 |
MXI1 |
MAX interactor 1, dimerization protein |
29175 |
0.18 |
| chr1_151445359_151445526 | 0.01 |
POGZ |
pogo transposable element with ZNF domain |
13501 |
0.11 |
| chr22_29191548_29191699 | 0.01 |
XBP1 |
X-box binding protein 1 |
4498 |
0.15 |
| chr8_134501895_134502046 | 0.01 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
27 |
0.99 |
| chr19_19538149_19538407 | 0.01 |
GATAD2A |
GATA zinc finger domain containing 2A |
47 |
0.96 |
| chr5_130870514_130870903 | 0.01 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
1982 |
0.48 |
| chr16_17091340_17091491 | 0.01 |
CTD-2576D5.4 |
|
136946 |
0.05 |
| chr17_66242892_66243078 | 0.01 |
AMZ2 |
archaelysin family metallopeptidase 2 |
730 |
0.5 |
| chr22_44207665_44207833 | 0.01 |
EFCAB6 |
EF-hand calcium binding domain 6 |
321 |
0.69 |
| chr19_16205574_16205925 | 0.01 |
TPM4 |
tropomyosin 4 |
1367 |
0.36 |
| chr5_102729544_102729916 | 0.01 |
C5orf30 |
chromosome 5 open reading frame 30 |
134605 |
0.05 |
| chr22_46025392_46025543 | 0.01 |
ENSG00000251985 |
. |
4955 |
0.22 |
| chr3_32445320_32445471 | 0.01 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
11864 |
0.25 |
| chrX_39169100_39169251 | 0.00 |
ENSG00000207122 |
. |
228637 |
0.02 |
| chr9_79218995_79219152 | 0.00 |
ENSG00000241781 |
. |
32342 |
0.18 |
| chr10_34411268_34411541 | 0.00 |
ENSG00000199200 |
. |
79598 |
0.12 |
| chr1_32389459_32389792 | 0.00 |
PTP4A2 |
protein tyrosine phosphatase type IVA, member 2 |
4541 |
0.2 |
| chr7_43641940_43642091 | 0.00 |
STK17A |
serine/threonine kinase 17a |
19351 |
0.17 |
| chr11_118755028_118755179 | 0.00 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
628 |
0.54 |
| chr5_68660810_68660961 | 0.00 |
TAF9 |
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
1763 |
0.28 |
| chr16_2178397_2178612 | 0.00 |
ENSG00000200059 |
. |
3348 |
0.07 |
| chr22_40768850_40769001 | 0.00 |
SGSM3 |
small G protein signaling modulator 3 |
2290 |
0.25 |
| chr2_152220395_152220546 | 0.00 |
ENSG00000264684 |
. |
4378 |
0.19 |
| chr8_74889988_74890139 | 0.00 |
TMEM70 |
transmembrane protein 70 |
1619 |
0.27 |
| chr9_83990440_83990591 | 0.00 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
237922 |
0.02 |
| chr3_158288758_158289050 | 0.00 |
MLF1 |
myeloid leukemia factor 1 |
48 |
0.5 |
| chr15_33122968_33123119 | 0.00 |
FMN1 |
formin 1 |
57412 |
0.12 |
| chr6_30118578_30118729 | 0.00 |
TRIM10 |
tripartite motif containing 10 |
10058 |
0.1 |
| chr2_235383990_235384141 | 0.00 |
ARL4C |
ADP-ribosylation factor-like 4C |
21179 |
0.29 |
| chr20_42575036_42575187 | 0.00 |
TOX2 |
TOX high mobility group box family member 2 |
766 |
0.73 |
| chr2_28114153_28114304 | 0.00 |
RBKS |
ribokinase |
263 |
0.74 |
| chr2_43036312_43036514 | 0.00 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
16681 |
0.23 |
| chr12_27939071_27939324 | 0.00 |
RP11-860B13.1 |
|
5197 |
0.13 |
| chr3_115231996_115232281 | 0.00 |
GAP43 |
growth associated protein 43 |
110033 |
0.07 |
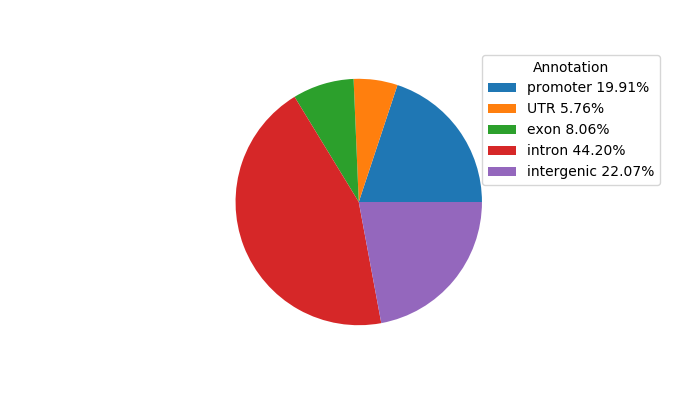
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |