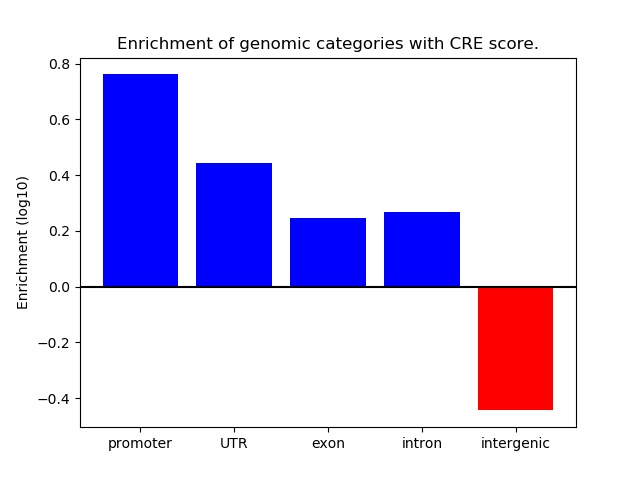
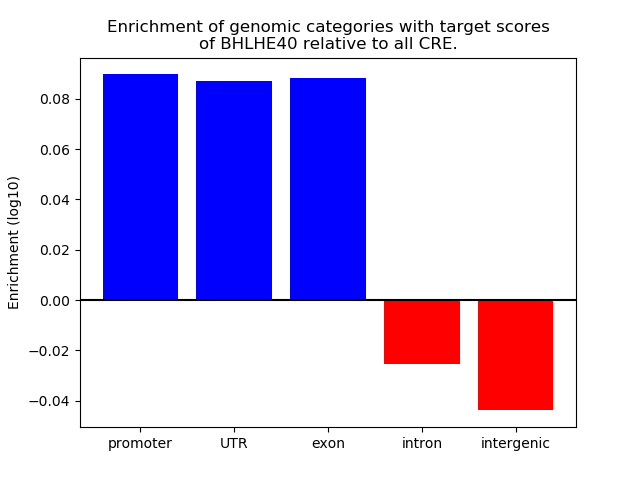
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
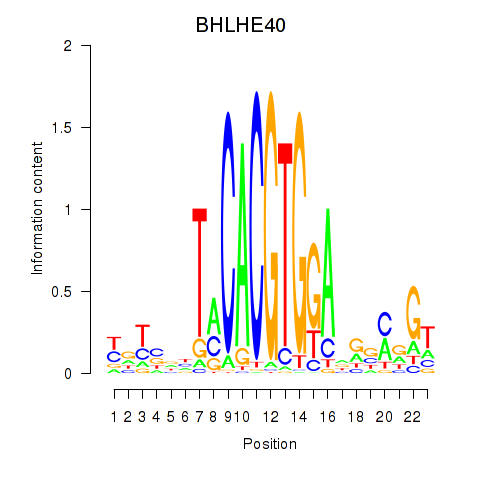
Results for BHLHE40
Z-value: 1.28
Transcription factors associated with BHLHE40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE40
|
ENSG00000134107.4 | basic helix-loop-helix family member e40 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_4991634_4991797 | BHLHE40 | 29086 | 0.162405 | -0.53 | 1.4e-01 | Click! |
| chr3_5020639_5020790 | BHLHE40 | 87 | 0.941773 | 0.51 | 1.6e-01 | Click! |
| chr3_4994783_4994934 | BHLHE40 | 25943 | 0.169522 | -0.49 | 1.8e-01 | Click! |
| chr3_5019096_5019422 | BHLHE40 | 1542 | 0.374689 | 0.41 | 2.7e-01 | Click! |
| chr3_5018310_5018816 | BHLHE40 | 2238 | 0.297469 | 0.37 | 3.3e-01 | Click! |
Activity of the BHLHE40 motif across conditions
Conditions sorted by the z-value of the BHLHE40 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
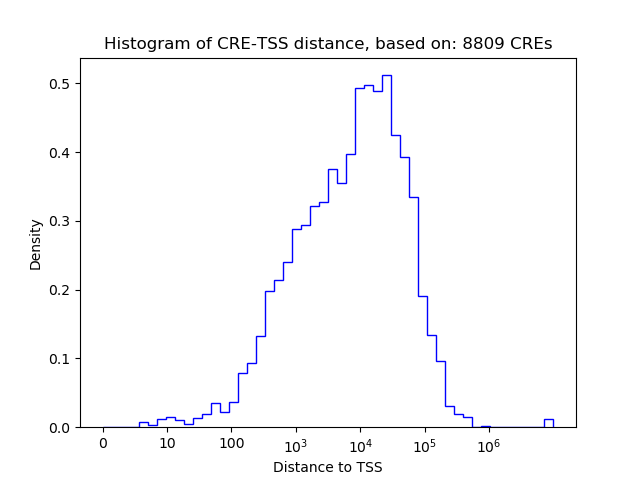
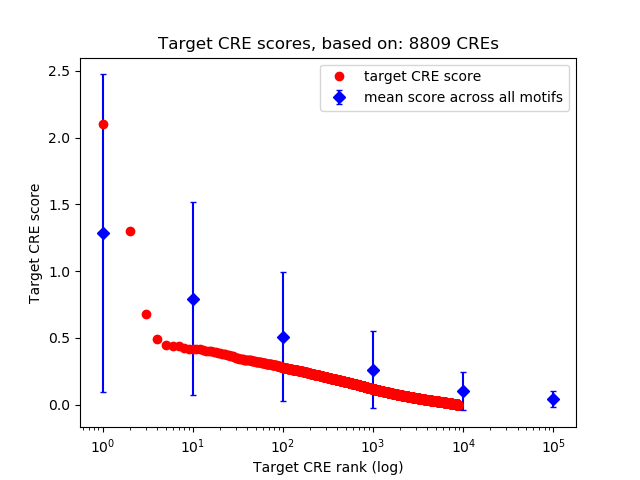
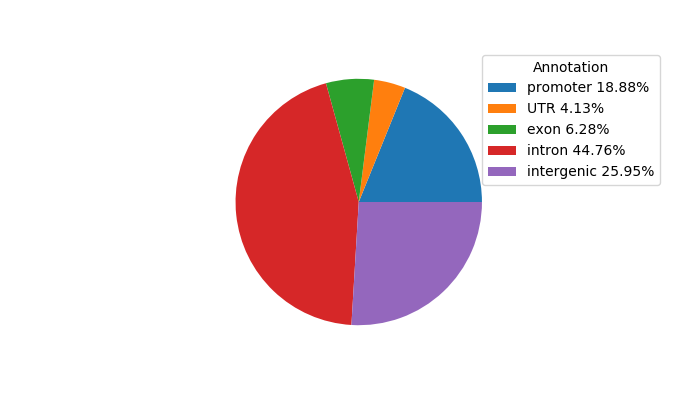
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_157548637_157548975 | 2.10 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
78761 |
0.11 |
| chr6_157548368_157548619 | 1.30 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
78448 |
0.11 |
| chr6_150401228_150401379 | 0.68 |
ULBP3 |
UL16 binding protein 3 |
11072 |
0.18 |
| chr11_95393873_95394024 | 0.49 |
FAM76B |
family with sequence similarity 76, member B |
126021 |
0.05 |
| chr4_40303302_40303674 | 0.45 |
CHRNA9 |
cholinergic receptor, nicotinic, alpha 9 (neuronal) |
33858 |
0.17 |
| chr1_209944656_209944807 | 0.44 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
2771 |
0.19 |
| chr7_2070925_2071076 | 0.44 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
48648 |
0.17 |
| chr9_92138368_92138605 | 0.42 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
25441 |
0.19 |
| chr11_118178245_118178503 | 0.42 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
2760 |
0.18 |
| chr2_106604937_106605088 | 0.42 |
C2orf40 |
chromosome 2 open reading frame 40 |
74738 |
0.11 |
| chr15_93579130_93579323 | 0.42 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
27130 |
0.2 |
| chr20_57973693_57973970 | 0.42 |
ENSG00000263903 |
. |
52296 |
0.16 |
| chr1_42271419_42271644 | 0.41 |
ENSG00000264896 |
. |
46719 |
0.18 |
| chr2_149284716_149284924 | 0.40 |
MBD5 |
methyl-CpG binding domain protein 5 |
58526 |
0.15 |
| chr1_2950991_2951142 | 0.40 |
ACTRT2 |
actin-related protein T2 |
13020 |
0.19 |
| chr9_92105986_92106169 | 0.40 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
6270 |
0.24 |
| chr3_46416627_46416863 | 0.40 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
2584 |
0.23 |
| chr6_71360263_71360634 | 0.39 |
SMAP1 |
small ArfGAP 1 |
17031 |
0.26 |
| chr3_128032806_128032957 | 0.39 |
ENSG00000221067 |
. |
48127 |
0.15 |
| chr7_150639030_150639181 | 0.38 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
13889 |
0.15 |
| chr2_169290603_169290754 | 0.38 |
ENSG00000239230 |
. |
7037 |
0.21 |
| chr12_58153080_58153231 | 0.38 |
MARCH9 |
membrane-associated ring finger (C3HC4) 9 |
1885 |
0.12 |
| chr12_25432795_25432959 | 0.38 |
KRAS |
Kirsten rat sarcoma viral oncogene homolog |
29007 |
0.18 |
| chr20_50720819_50720982 | 0.37 |
ZFP64 |
ZFP64 zinc finger protein |
489 |
0.89 |
| chr1_241839762_241839978 | 0.37 |
WDR64 |
WD repeat domain 64 |
7012 |
0.22 |
| chr1_235377409_235377560 | 0.37 |
ARID4B |
AT rich interactive domain 4B (RBP1-like) |
253 |
0.92 |
| chr7_1085411_1085702 | 0.37 |
GPR146 |
G protein-coupled receptor 146 |
1344 |
0.26 |
| chr14_100540531_100540793 | 0.36 |
CTD-2376I20.1 |
|
553 |
0.71 |
| chr7_139924008_139924237 | 0.36 |
ENSG00000199971 |
. |
14651 |
0.16 |
| chr11_748009_748200 | 0.35 |
TALDO1 |
transaldolase 1 |
676 |
0.44 |
| chr15_44958521_44958824 | 0.35 |
PATL2 |
protein associated with topoisomerase II homolog 2 (yeast) |
2011 |
0.24 |
| chr13_43563311_43563462 | 0.35 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
1981 |
0.43 |
| chr2_113943965_113944116 | 0.35 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
9817 |
0.14 |
| chr9_123641457_123641690 | 0.35 |
PHF19 |
PHD finger protein 19 |
1967 |
0.31 |
| chr16_89850957_89851182 | 0.34 |
FANCA |
Fanconi anemia, complementation group A |
31974 |
0.1 |
| chr3_18416180_18416331 | 0.34 |
RP11-158G18.1 |
|
34872 |
0.19 |
| chr10_63810330_63810481 | 0.34 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
1435 |
0.54 |
| chr15_65075233_65075384 | 0.34 |
RBPMS2 |
RNA binding protein with multiple splicing 2 |
7522 |
0.14 |
| chr15_32635679_32635830 | 0.34 |
ENSG00000221444 |
. |
49919 |
0.09 |
| chr1_101742974_101743225 | 0.34 |
RP4-575N6.5 |
|
34385 |
0.14 |
| chr19_16486076_16486459 | 0.34 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
13503 |
0.14 |
| chr7_50426987_50427896 | 0.33 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
60196 |
0.13 |
| chr5_131347971_131348122 | 0.33 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
176 |
0.88 |
| chr6_36487605_36487756 | 0.33 |
ENSG00000239964 |
. |
2288 |
0.23 |
| chr1_194025907_194026313 | 0.33 |
ENSG00000252241 |
. |
325036 |
0.01 |
| chrX_128889315_128889636 | 0.33 |
XPNPEP2 |
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
16477 |
0.19 |
| chr9_4605304_4605466 | 0.33 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
33050 |
0.16 |
| chr16_57041025_57041193 | 0.33 |
NLRC5 |
NLR family, CARD domain containing 5 |
2711 |
0.2 |
| chr1_209944824_209944985 | 0.33 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
2944 |
0.19 |
| chr17_38076794_38077078 | 0.32 |
GSDMB |
gasdermin B |
2033 |
0.21 |
| chr12_2799901_2800122 | 0.32 |
CACNA1C-AS1 |
CACNA1C antisense RNA 1 |
273 |
0.91 |
| chr3_33101505_33101658 | 0.32 |
GLB1 |
galactosidase, beta 1 |
36703 |
0.13 |
| chr12_92726454_92726695 | 0.32 |
RP11-693J15.4 |
|
88733 |
0.08 |
| chr1_12149159_12149332 | 0.32 |
ENSG00000201135 |
. |
11204 |
0.15 |
| chr17_2699183_2699336 | 0.32 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
473 |
0.8 |
| chr14_25162667_25162947 | 0.32 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
59334 |
0.11 |
| chr17_18858474_18858633 | 0.32 |
AC090286.4 |
|
1091 |
0.36 |
| chr16_57634856_57635007 | 0.31 |
GPR56 |
G protein-coupled receptor 56 |
9633 |
0.15 |
| chr22_50754075_50754226 | 0.31 |
XX-C283C717.1 |
|
1090 |
0.31 |
| chr5_157940155_157940453 | 0.31 |
CTD-2363C16.1 |
|
469710 |
0.01 |
| chr9_135382524_135382903 | 0.31 |
BARHL1 |
BarH-like homeobox 1 |
74859 |
0.09 |
| chr11_64821935_64822086 | 0.31 |
NAALADL1 |
N-acetylated alpha-linked acidic dipeptidase-like 1 |
3983 |
0.08 |
| chr5_171543346_171543497 | 0.31 |
ENSG00000266671 |
. |
35441 |
0.16 |
| chr1_112045793_112046152 | 0.31 |
ADORA3 |
adenosine A3 receptor |
138 |
0.93 |
| chr7_132121999_132122205 | 0.31 |
AC011625.1 |
|
85009 |
0.1 |
| chr2_10815541_10815692 | 0.31 |
NOL10 |
nucleolar protein 10 |
3800 |
0.22 |
| chr7_158991213_158991434 | 0.31 |
VIPR2 |
vasoactive intestinal peptide receptor 2 |
53674 |
0.18 |
| chr12_131796444_131796595 | 0.31 |
ENSG00000212251 |
. |
12223 |
0.23 |
| chr3_187692256_187692598 | 0.31 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
178645 |
0.03 |
| chr10_11309961_11310112 | 0.30 |
CELF2-AS1 |
CELF2 antisense RNA 1 |
51811 |
0.15 |
| chr5_98267544_98267695 | 0.30 |
ENSG00000200351 |
. |
4832 |
0.23 |
| chr3_45906834_45907101 | 0.30 |
CCR9 |
chemokine (C-C motif) receptor 9 |
21029 |
0.15 |
| chr2_198143456_198143859 | 0.30 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
23586 |
0.16 |
| chr20_62859464_62859615 | 0.30 |
PCMTD2 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
27555 |
0.17 |
| chr5_1313612_1313923 | 0.30 |
ENSG00000263670 |
. |
4275 |
0.2 |
| chr1_20013079_20013230 | 0.30 |
HTR6 |
5-hydroxytryptamine (serotonin) receptor 6, G protein-coupled |
21374 |
0.16 |
| chr11_114053249_114053976 | 0.30 |
NNMT |
nicotinamide N-methyltransferase |
74941 |
0.09 |
| chr11_96024786_96024937 | 0.30 |
ENSG00000266192 |
. |
49741 |
0.14 |
| chr17_73239171_73239322 | 0.30 |
GGA3 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
4084 |
0.11 |
| chr11_113955241_113955493 | 0.30 |
ENSG00000221112 |
. |
2284 |
0.34 |
| chr18_21509698_21509962 | 0.30 |
LAMA3 |
laminin, alpha 3 |
1627 |
0.41 |
| chr14_98899836_98899987 | 0.30 |
ENSG00000241757 |
. |
81910 |
0.11 |
| chr2_128399438_128399670 | 0.30 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
152 |
0.94 |
| chr15_61135667_61135839 | 0.29 |
RP11-554D20.1 |
|
78814 |
0.11 |
| chr17_76745970_76746190 | 0.29 |
CYTH1 |
cytohesin 1 |
13108 |
0.19 |
| chr6_137710654_137710959 | 0.29 |
OLIG3 |
oligodendrocyte transcription factor 3 |
104725 |
0.07 |
| chr1_227949174_227949396 | 0.29 |
SNAP47 |
synaptosomal-associated protein, 47kDa |
13521 |
0.16 |
| chr8_100024823_100024974 | 0.29 |
VPS13B |
vacuolar protein sorting 13 homolog B (yeast) |
596 |
0.74 |
| chr1_160645057_160645403 | 0.29 |
RP11-404F10.2 |
|
1918 |
0.29 |
| chr21_44252788_44252947 | 0.29 |
WDR4 |
WD repeat domain 4 |
46774 |
0.13 |
| chr5_133376557_133376708 | 0.29 |
VDAC1 |
voltage-dependent anion channel 1 |
35808 |
0.17 |
| chr19_42027703_42027944 | 0.29 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
28063 |
0.13 |
| chr2_106404052_106404457 | 0.29 |
NCK2 |
NCK adaptor protein 2 |
28761 |
0.23 |
| chr14_100537205_100537447 | 0.28 |
CTD-2376I20.1 |
|
3889 |
0.18 |
| chr12_133349779_133349930 | 0.28 |
ANKLE2 |
ankyrin repeat and LEM domain containing 2 |
11380 |
0.15 |
| chr2_662119_662318 | 0.28 |
TMEM18 |
transmembrane protein 18 |
13557 |
0.2 |
| chr8_27226083_27226368 | 0.28 |
PTK2B |
protein tyrosine kinase 2 beta |
11943 |
0.22 |
| chr1_229228666_229228817 | 0.28 |
RP5-1061H20.5 |
|
134568 |
0.04 |
| chr9_140165488_140165705 | 0.28 |
TOR4A |
torsin family 4, member A |
6605 |
0.07 |
| chr10_126849895_126850046 | 0.28 |
CTBP2 |
C-terminal binding protein 2 |
340 |
0.93 |
| chrX_46471596_46471846 | 0.28 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
38502 |
0.17 |
| chr1_54815665_54815816 | 0.28 |
SSBP3 |
single stranded DNA binding protein 3 |
55437 |
0.11 |
| chr13_114906998_114907153 | 0.28 |
RASA3 |
RAS p21 protein activator 3 |
8989 |
0.22 |
| chr8_66854346_66854497 | 0.27 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
79374 |
0.1 |
| chr17_78637163_78637475 | 0.27 |
RPTOR |
regulatory associated protein of MTOR, complex 1 |
118051 |
0.05 |
| chr1_1107776_1107927 | 0.27 |
TTLL10 |
tubulin tyrosine ligase-like family, member 10 |
1432 |
0.2 |
| chr17_71826931_71827082 | 0.27 |
SDK2 |
sidekick cell adhesion molecule 2 |
186778 |
0.03 |
| chr14_22409773_22409924 | 0.27 |
ENSG00000222776 |
. |
161063 |
0.03 |
| chr15_64151124_64151275 | 0.27 |
ENSG00000199156 |
. |
12019 |
0.21 |
| chr16_27206515_27206666 | 0.27 |
KDM8 |
lysine (K)-specific demethylase 8 |
8217 |
0.19 |
| chr4_89202595_89202746 | 0.27 |
PPM1K |
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
2372 |
0.23 |
| chr21_46496959_46497110 | 0.27 |
ADARB1 |
adenosine deaminase, RNA-specific, B1 |
2519 |
0.22 |
| chr20_47430608_47430759 | 0.27 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
13737 |
0.26 |
| chr14_61827530_61827767 | 0.27 |
PRKCH |
protein kinase C, eta |
144 |
0.97 |
| chr6_167524624_167524999 | 0.27 |
CCR6 |
chemokine (C-C motif) receptor 6 |
484 |
0.81 |
| chr13_99849164_99849315 | 0.27 |
UBAC2 |
UBA domain containing 2 |
3789 |
0.23 |
| chr16_85933118_85933269 | 0.27 |
IRF8 |
interferon regulatory factor 8 |
424 |
0.87 |
| chr14_105532821_105533069 | 0.27 |
GPR132 |
G protein-coupled receptor 132 |
1163 |
0.49 |
| chr7_2750642_2751031 | 0.27 |
AMZ1 |
archaelysin family metallopeptidase 1 |
23000 |
0.18 |
| chr2_3243298_3243449 | 0.27 |
TSSC1-IT1 |
TSSC1 intronic transcript 1 (non-protein coding) |
61863 |
0.13 |
| chr5_56122812_56123396 | 0.27 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
11703 |
0.17 |
| chr9_92450655_92450815 | 0.27 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
230782 |
0.02 |
| chr10_22969605_22969760 | 0.27 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
33355 |
0.22 |
| chr8_125685655_125685918 | 0.27 |
MTSS1 |
metastasis suppressor 1 |
54071 |
0.13 |
| chr21_32929274_32929425 | 0.27 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
1941 |
0.32 |
| chr10_8087441_8087780 | 0.27 |
GATA3 |
GATA binding protein 3 |
9046 |
0.32 |
| chr12_112545861_112546012 | 0.26 |
NAA25 |
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
611 |
0.66 |
| chr1_182599288_182599608 | 0.26 |
RGS16 |
regulator of G-protein signaling 16 |
25905 |
0.17 |
| chr6_157548074_157548363 | 0.26 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
78173 |
0.11 |
| chr19_15624982_15625133 | 0.26 |
CYP4F22 |
cytochrome P450, family 4, subfamily F, polypeptide 22 |
5753 |
0.16 |
| chr17_43307469_43307750 | 0.26 |
CTD-2020K17.1 |
|
8020 |
0.1 |
| chr3_128518442_128518718 | 0.26 |
RAB7A |
RAB7A, member RAS oncogene family |
4378 |
0.21 |
| chr2_106371801_106372035 | 0.26 |
NCK2 |
NCK adaptor protein 2 |
9730 |
0.3 |
| chr1_157693341_157693492 | 0.26 |
FCRL3 |
Fc receptor-like 3 |
22769 |
0.17 |
| chr3_171826118_171826452 | 0.26 |
FNDC3B |
fibronectin type III domain containing 3B |
18479 |
0.26 |
| chr20_62362374_62362787 | 0.26 |
LIME1 |
Lck interacting transmembrane adaptor 1 |
5055 |
0.09 |
| chr21_45502719_45503242 | 0.26 |
PWP2 |
PWP2 periodic tryptophan protein homolog (yeast) |
24191 |
0.15 |
| chr16_68389761_68390217 | 0.26 |
SMPD3 |
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
14919 |
0.11 |
| chr4_2816087_2816379 | 0.26 |
SH3BP2 |
SH3-domain binding protein 2 |
2202 |
0.32 |
| chr3_186760165_186760316 | 0.26 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
1291 |
0.54 |
| chr1_226039631_226039809 | 0.26 |
RP11-285F7.2 |
|
5497 |
0.14 |
| chr12_133014236_133014611 | 0.26 |
MUC8 |
mucin 8 |
36303 |
0.16 |
| chr20_57735532_57735874 | 0.26 |
ZNF831 |
zinc finger protein 831 |
30372 |
0.2 |
| chr1_42269178_42269372 | 0.26 |
ENSG00000264896 |
. |
44463 |
0.19 |
| chr6_167460456_167460849 | 0.26 |
FGFR1OP |
FGFR1 oncogene partner |
47756 |
0.1 |
| chr8_144910846_144910997 | 0.26 |
PUF60 |
poly-U binding splicing factor 60KDa |
275 |
0.81 |
| chr7_152039023_152039174 | 0.26 |
ENSG00000253088 |
. |
60767 |
0.12 |
| chr16_17440476_17440787 | 0.26 |
XYLT1 |
xylosyltransferase I |
124107 |
0.06 |
| chr15_76574850_76575217 | 0.26 |
ETFA |
electron-transfer-flavoprotein, alpha polypeptide |
9799 |
0.23 |
| chr20_60867419_60867570 | 0.25 |
ADRM1 |
adhesion regulating molecule 1 |
10567 |
0.12 |
| chr18_21551706_21552127 | 0.25 |
TTC39C |
tetratricopeptide repeat domain 39C |
20821 |
0.16 |
| chr10_126414122_126414344 | 0.25 |
FAM53B |
family with sequence similarity 53, member B |
17306 |
0.16 |
| chr21_43834242_43834420 | 0.25 |
ENSG00000252619 |
. |
3371 |
0.17 |
| chr22_37641156_37641409 | 0.25 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
794 |
0.57 |
| chr13_100039907_100040178 | 0.25 |
ENSG00000207719 |
. |
31657 |
0.16 |
| chr1_78354309_78354460 | 0.25 |
NEXN |
nexilin (F actin binding protein) |
37 |
0.95 |
| chr3_187719085_187719236 | 0.25 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
151912 |
0.04 |
| chr11_76376085_76376254 | 0.25 |
LRRC32 |
leucine rich repeat containing 32 |
4875 |
0.2 |
| chr1_101711671_101711822 | 0.25 |
RP4-575N6.5 |
|
3032 |
0.21 |
| chr1_77884409_77884560 | 0.25 |
ENSG00000251767 |
. |
1526 |
0.44 |
| chr13_110222272_110222771 | 0.25 |
LINC00676 |
long intergenic non-protein coding RNA 676 |
158108 |
0.04 |
| chr7_31064429_31064580 | 0.25 |
ADCYAP1R1 |
adenylate cyclase activating polypeptide 1 (pituitary) receptor type I |
27572 |
0.21 |
| chr2_64553101_64553267 | 0.25 |
ENSG00000264297 |
. |
14709 |
0.25 |
| chr13_114909698_114910252 | 0.25 |
RASA3 |
RAS p21 protein activator 3 |
11889 |
0.21 |
| chr1_200980853_200981004 | 0.25 |
KIF21B |
kinesin family member 21B |
11608 |
0.18 |
| chr3_38036912_38037201 | 0.25 |
VILL |
villin-like |
1412 |
0.32 |
| chr6_20206050_20206262 | 0.25 |
RP11-239H6.2 |
|
6162 |
0.24 |
| chr11_2489014_2489234 | 0.25 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
6007 |
0.18 |
| chr10_13932808_13933126 | 0.25 |
FRMD4A |
FERM domain containing 4A |
32055 |
0.2 |
| chr14_100610702_100610964 | 0.25 |
DEGS2 |
delta(4)-desaturase, sphingolipid 2 |
15099 |
0.12 |
| chr22_26690400_26690699 | 0.25 |
SEZ6L |
seizure related 6 homolog (mouse)-like |
2332 |
0.34 |
| chr7_92666364_92666515 | 0.25 |
ENSG00000266794 |
. |
65826 |
0.11 |
| chr3_9992673_9992875 | 0.25 |
PRRT3 |
proline-rich transmembrane protein 3 |
1300 |
0.22 |
| chr15_34695379_34695530 | 0.25 |
GOLGA8A |
golgin A8 family, member A |
4434 |
0.16 |
| chr11_14076377_14076528 | 0.25 |
ENSG00000212365 |
. |
80080 |
0.11 |
| chr4_100739674_100739989 | 0.25 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
1828 |
0.43 |
| chr2_219233652_219233882 | 0.24 |
ENSG00000225062 |
. |
1209 |
0.27 |
| chr16_50318201_50318668 | 0.24 |
ADCY7 |
adenylate cyclase 7 |
3388 |
0.25 |
| chr18_21547738_21547925 | 0.24 |
LAMA3 |
laminin, alpha 3 |
17974 |
0.17 |
| chr7_2138652_2138817 | 0.24 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
8609 |
0.28 |
| chr10_102107561_102107838 | 0.24 |
SCD |
stearoyl-CoA desaturase (delta-9-desaturase) |
818 |
0.47 |
| chr15_53075384_53075631 | 0.24 |
ONECUT1 |
one cut homeobox 1 |
2127 |
0.43 |
| chr1_167618740_167619238 | 0.24 |
RP3-455J7.4 |
|
19078 |
0.18 |
| chr1_19400551_19400797 | 0.24 |
UBR4 |
ubiquitin protein ligase E3 component n-recognin 4 |
8060 |
0.2 |
| chr6_159462884_159463035 | 0.24 |
TAGAP |
T-cell activation RhoGTPase activating protein |
3091 |
0.25 |
| chr1_36172716_36172867 | 0.24 |
ENSG00000239859 |
. |
916 |
0.53 |
| chrX_120693101_120693333 | 0.24 |
ENSG00000265456 |
. |
188391 |
0.03 |
| chr16_27244955_27245106 | 0.24 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
631 |
0.7 |
| chr3_59430359_59430723 | 0.24 |
C3orf67 |
chromosome 3 open reading frame 67 |
394731 |
0.01 |
| chr12_9148612_9148857 | 0.24 |
RP11-259O18.4 |
|
428 |
0.8 |
| chr19_30166083_30166320 | 0.24 |
PLEKHF1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
8420 |
0.23 |
| chr3_28294093_28294244 | 0.24 |
CMC1 |
C-x(9)-C motif containing 1 |
1051 |
0.66 |
| chr15_34841604_34841755 | 0.24 |
GOLGA8B |
golgin A8 family, member B |
13468 |
0.17 |
| chr9_96357787_96357938 | 0.24 |
PHF2 |
PHD finger protein 2 |
18953 |
0.19 |
| chr13_99214255_99214406 | 0.24 |
STK24 |
serine/threonine kinase 24 |
14787 |
0.2 |
| chr8_11309548_11309973 | 0.24 |
FAM167A |
family with sequence similarity 167, member A |
6882 |
0.16 |
| chr15_81299308_81299459 | 0.24 |
C15orf26 |
chromosome 15 open reading frame 26 |
9 |
0.97 |
| chr8_61694776_61694927 | 0.24 |
RP11-33I11.2 |
|
27314 |
0.22 |
| chr12_53593792_53594465 | 0.23 |
ITGB7 |
integrin, beta 7 |
99 |
0.94 |
| chr3_71187126_71187427 | 0.23 |
FOXP1 |
forkhead box P1 |
7288 |
0.32 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.4 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.3 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.1 | 0.2 | GO:0002249 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.2 | GO:0007132 | meiotic metaphase I(GO:0007132) metaphase(GO:0051323) |
| 0.1 | 0.3 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) positive regulation of cardiocyte differentiation(GO:1905209) |
| 0.1 | 0.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0002666 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.2 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0002335 | mature B cell differentiation involved in immune response(GO:0002313) mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.2 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) |
| 0.0 | 0.1 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.6 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.3 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0003077 | obsolete negative regulation of diuresis(GO:0003077) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.2 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0070100 | regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.0 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0090382 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:1901985 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.4 | GO:0031670 | cellular response to nutrient(GO:0031670) |
| 0.0 | 0.2 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.2 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.1 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0052255 | modulation by symbiont of host defense response(GO:0052031) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0032366 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.4 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.0 | 0.1 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.0 | GO:0072577 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.1 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0044266 | multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.0 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.2 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.1 | GO:0071501 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.5 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.2 | GO:0050927 | positive regulation of positive chemotaxis(GO:0050927) |
| 0.0 | 0.2 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.0 | GO:2000318 | T-helper cell lineage commitment(GO:0002295) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:0045910 | negative regulation of DNA recombination(GO:0045910) |
| 0.0 | 0.0 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.0 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.2 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.0 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.0 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.0 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.0 | GO:0060872 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.0 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.3 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.1 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.4 | GO:0019079 | viral genome replication(GO:0019079) |
| 0.0 | 0.1 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.0 | 0.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.3 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.0 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.0 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) |
| 0.0 | 0.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0032222 | regulation of synaptic transmission, cholinergic(GO:0032222) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.0 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.0 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.1 | GO:0051806 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.2 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.1 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0051000 | positive regulation of nitric-oxide synthase activity(GO:0051000) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:1903319 | positive regulation of protein processing(GO:0010954) positive regulation of protein maturation(GO:1903319) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.0 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.0 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.0 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.0 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0045581 | negative regulation of T cell differentiation(GO:0045581) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0071600 | otic vesicle formation(GO:0030916) otic vesicle development(GO:0071599) otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.0 | GO:0072124 | cellular response to heparin(GO:0071504) glomerular mesangial cell proliferation(GO:0072110) regulation of glomerular mesangial cell proliferation(GO:0072124) metanephric glomerulus development(GO:0072224) metanephric glomerulus vasculature development(GO:0072239) regulation of glomerulus development(GO:0090192) |
| 0.0 | 0.0 | GO:0046636 | negative regulation of alpha-beta T cell activation(GO:0046636) |
| 0.0 | 0.0 | GO:0015919 | peroxisomal membrane transport(GO:0015919) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0031054 | pre-miRNA processing(GO:0031054) |
| 0.0 | 0.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0098573 | intrinsic component of mitochondrial membrane(GO:0098573) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.0 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0032142 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.0 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.5 | GO:0051540 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.0 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.4 | REACTOME CELL CYCLE CHECKPOINTS | Genes involved in Cell Cycle Checkpoints |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |