Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
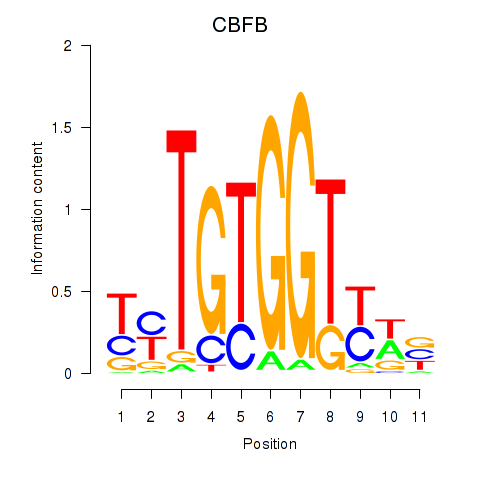
Results for CBFB
Z-value: 0.64
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.9 | core-binding factor subunit beta |
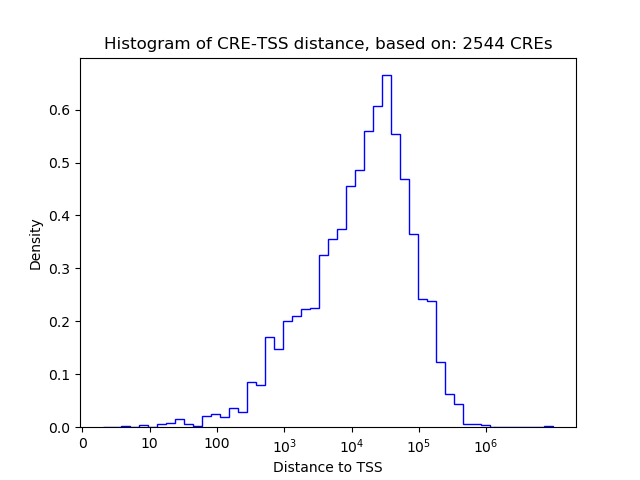
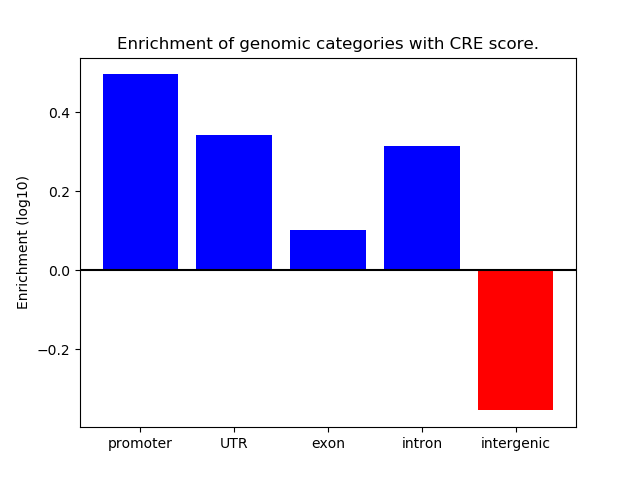
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_67096782_67097112 | CBFB | 33092 | 0.087772 | -0.90 | 1.1e-03 | Click! |
| chr16_67069269_67069524 | CBFB | 5541 | 0.131957 | -0.83 | 5.1e-03 | Click! |
| chr16_67068602_67069010 | CBFB | 4951 | 0.135685 | -0.77 | 1.6e-02 | Click! |
| chr16_67062545_67062763 | CBFB | 365 | 0.795605 | 0.67 | 4.7e-02 | Click! |
| chr16_67090389_67090540 | CBFB | 26609 | 0.098058 | 0.65 | 5.8e-02 | Click! |
Activity of the CBFB motif across conditions
Conditions sorted by the z-value of the CBFB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
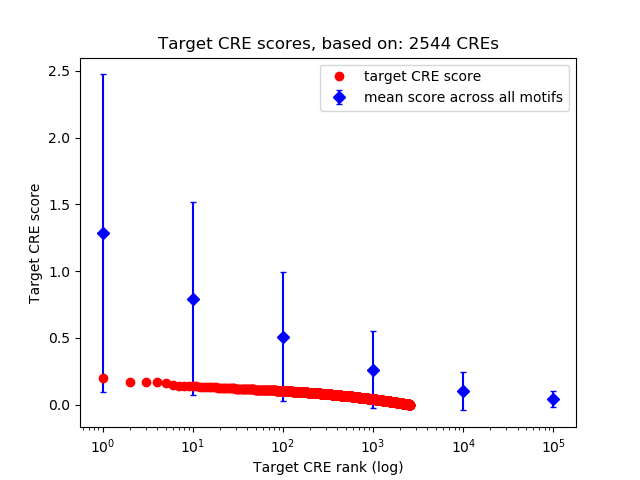
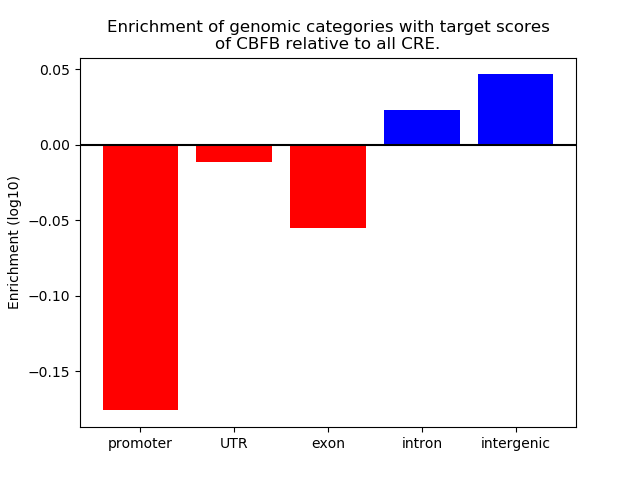
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_20877296_20877733 | 0.20 |
RP3-348I23.2 |
|
76589 |
0.12 |
| chr7_116417479_116417780 | 0.17 |
MET |
met proto-oncogene |
400 |
0.88 |
| chr1_16277969_16279055 | 0.17 |
ZBTB17 |
zinc finger and BTB domain containing 17 |
7589 |
0.15 |
| chr3_183563662_183564003 | 0.17 |
PARL |
presenilin associated, rhomboid-like |
1770 |
0.27 |
| chr13_80509988_80510139 | 0.16 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
403731 |
0.01 |
| chr1_201689199_201689500 | 0.15 |
ENSG00000264802 |
. |
713 |
0.58 |
| chr20_44407057_44407418 | 0.14 |
ENSG00000221046 |
. |
100 |
0.93 |
| chr1_214618510_214618915 | 0.14 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
19434 |
0.26 |
| chr2_20763043_20763331 | 0.14 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
29121 |
0.18 |
| chr7_78719932_78720083 | 0.14 |
ENSG00000212545 |
. |
60451 |
0.13 |
| chr6_135148528_135148679 | 0.14 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
101696 |
0.07 |
| chr15_67094989_67095410 | 0.14 |
SMAD6 |
SMAD family member 6 |
91164 |
0.08 |
| chr3_191094193_191094344 | 0.14 |
UTS2B |
urotensin 2B |
45943 |
0.15 |
| chr13_107152902_107153053 | 0.14 |
EFNB2 |
ephrin-B2 |
34485 |
0.22 |
| chr7_55176177_55176328 | 0.13 |
EGFR |
epidermal growth factor receptor |
1164 |
0.63 |
| chr18_77722000_77722505 | 0.13 |
HSBP1L1 |
heat shock factor binding protein 1-like 1 |
2309 |
0.28 |
| chr11_95239026_95239264 | 0.13 |
ENSG00000201204 |
. |
32607 |
0.24 |
| chr2_159902692_159902978 | 0.13 |
ENSG00000202029 |
. |
19181 |
0.24 |
| chr1_16126273_16126474 | 0.13 |
FBLIM1 |
filamin binding LIM protein 1 |
34925 |
0.09 |
| chr2_1593137_1593288 | 0.13 |
AC144450.1 |
|
30673 |
0.22 |
| chr6_20878021_20878263 | 0.13 |
RP3-348I23.2 |
|
77217 |
0.12 |
| chr15_71854429_71854724 | 0.13 |
THSD4 |
thrombospondin, type I, domain containing 4 |
15001 |
0.27 |
| chr15_39465381_39465532 | 0.13 |
RP11-624L4.1 |
|
54542 |
0.16 |
| chr17_2717970_2718121 | 0.12 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
18269 |
0.17 |
| chr22_30194899_30195349 | 0.12 |
ASCC2 |
activating signal cointegrator 1 complex subunit 2 |
2967 |
0.2 |
| chr3_168602559_168602710 | 0.12 |
MECOM |
MDS1 and EVI1 complex locus |
243188 |
0.02 |
| chr16_53809610_53810340 | 0.12 |
FTO |
fat mass and obesity associated |
71881 |
0.11 |
| chr9_111249128_111249373 | 0.12 |
ENSG00000222512 |
. |
128041 |
0.06 |
| chr7_75717509_75717660 | 0.12 |
ENSG00000251798 |
. |
822 |
0.54 |
| chr10_22723256_22723780 | 0.12 |
RP11-301N24.3 |
|
73417 |
0.1 |
| chr1_110419656_110419807 | 0.12 |
RP11-195M16.1 |
|
9155 |
0.18 |
| chr6_64450641_64450792 | 0.12 |
PHF3 |
PHD finger protein 3 |
34026 |
0.21 |
| chr18_43363110_43363261 | 0.12 |
RP11-116O18.3 |
|
36005 |
0.14 |
| chr12_53318601_53318752 | 0.12 |
KRT8 |
keratin 8 |
1577 |
0.3 |
| chr15_49966605_49966756 | 0.12 |
RP11-353B9.1 |
|
22344 |
0.2 |
| chr2_27984512_27984663 | 0.12 |
MRPL33 |
mitochondrial ribosomal protein L33 |
9997 |
0.16 |
| chr2_28032160_28032486 | 0.12 |
AC110084.1 |
|
23137 |
0.14 |
| chr2_20795415_20795848 | 0.12 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
3323 |
0.28 |
| chr11_61065672_61065835 | 0.12 |
VWCE |
von Willebrand factor C and EGF domains |
2857 |
0.17 |
| chr11_57542945_57543611 | 0.12 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
5797 |
0.16 |
| chr6_25229819_25229970 | 0.12 |
ENSG00000264238 |
. |
26413 |
0.14 |
| chr20_13312667_13312818 | 0.12 |
RP5-1077I2.3 |
|
86726 |
0.09 |
| chr17_60901995_60902146 | 0.12 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
16365 |
0.24 |
| chr9_112578402_112578553 | 0.12 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
35708 |
0.14 |
| chr1_214630285_214630560 | 0.12 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
7724 |
0.3 |
| chr8_23656477_23656628 | 0.12 |
ENSG00000207027 |
. |
7338 |
0.2 |
| chr1_23782598_23782924 | 0.12 |
ASAP3 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
19591 |
0.15 |
| chr11_65255454_65256217 | 0.11 |
AP000769.1 |
Uncharacterized protein |
33107 |
0.08 |
| chr17_17415621_17415852 | 0.11 |
RP11-524F11.1 |
|
5071 |
0.17 |
| chr20_7822496_7822647 | 0.11 |
HAO1 |
hydroxyacid oxidase (glycolate oxidase) 1 |
98550 |
0.08 |
| chr5_171840254_171840540 | 0.11 |
ENSG00000216127 |
. |
26361 |
0.19 |
| chr22_18515057_18515208 | 0.11 |
MICAL3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
7807 |
0.15 |
| chr21_39662347_39662498 | 0.11 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
6057 |
0.3 |
| chr18_42745920_42746113 | 0.11 |
RP11-846C15.2 |
|
15063 |
0.25 |
| chr8_132869584_132869735 | 0.11 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
46676 |
0.2 |
| chr9_118837376_118837527 | 0.11 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
78632 |
0.11 |
| chr1_56431807_56432104 | 0.11 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
27002 |
0.27 |
| chr8_119065439_119065590 | 0.11 |
EXT1 |
exostosin glycosyltransferase 1 |
57139 |
0.17 |
| chr8_78478320_78478688 | 0.11 |
ENSG00000222334 |
. |
279452 |
0.02 |
| chr2_20366966_20367125 | 0.11 |
ENSG00000266059 |
. |
8062 |
0.18 |
| chr10_69914310_69914657 | 0.11 |
ENSG00000222371 |
. |
3353 |
0.26 |
| chr13_109896290_109896441 | 0.11 |
MYO16-AS1 |
MYO16 antisense RNA 1 |
42534 |
0.21 |
| chr17_36611263_36611448 | 0.11 |
ARHGAP23 |
Rho GTPase activating protein 23 |
2289 |
0.24 |
| chr4_122589997_122590148 | 0.11 |
ANXA5 |
annexin A5 |
28045 |
0.21 |
| chr8_131248547_131249235 | 0.11 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
10397 |
0.28 |
| chr6_47209724_47210383 | 0.11 |
TNFRSF21 |
tumor necrosis factor receptor superfamily, member 21 |
67588 |
0.13 |
| chr5_138206308_138206459 | 0.11 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
3344 |
0.24 |
| chr11_75735939_75736309 | 0.11 |
UVRAG |
UV radiation resistance associated |
41644 |
0.15 |
| chr3_16024682_16024833 | 0.11 |
ENSG00000207815 |
. |
109479 |
0.06 |
| chr2_241260003_241260423 | 0.11 |
ENSG00000221412 |
. |
58936 |
0.12 |
| chr6_144585105_144585256 | 0.11 |
UTRN |
utrophin |
21657 |
0.24 |
| chr12_25017223_25017374 | 0.11 |
RP11-625L16.3 |
|
33943 |
0.15 |
| chr18_25423566_25423717 | 0.11 |
AC015933.2 |
|
110842 |
0.08 |
| chr18_77721545_77721696 | 0.11 |
HSBP1L1 |
heat shock factor binding protein 1-like 1 |
2941 |
0.24 |
| chr15_63189996_63190147 | 0.11 |
RP11-1069G10.1 |
|
13838 |
0.23 |
| chr7_112121608_112121759 | 0.11 |
LSMEM1 |
leucine-rich single-pass membrane protein 1 |
637 |
0.76 |
| chr2_235960046_235960269 | 0.11 |
SH3BP4 |
SH3-domain binding protein 4 |
56673 |
0.17 |
| chr10_60420713_60420913 | 0.11 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
132482 |
0.05 |
| chr8_28250988_28251498 | 0.11 |
ZNF395 |
zinc finger protein 395 |
7260 |
0.15 |
| chr6_112524793_112524944 | 0.11 |
ENSG00000207044 |
. |
7120 |
0.18 |
| chr12_52848518_52848669 | 0.11 |
KRT6B |
keratin 6B |
2683 |
0.15 |
| chr6_149081739_149081986 | 0.11 |
UST |
uronyl-2-sulfotransferase |
13398 |
0.3 |
| chr16_58685994_58686145 | 0.11 |
CNOT1 |
CCR4-NOT transcription complex, subunit 1 |
22279 |
0.14 |
| chr5_97967989_97968140 | 0.11 |
ENSG00000223053 |
. |
6871 |
0.32 |
| chr7_2155638_2155836 | 0.11 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
6501 |
0.29 |
| chr6_169435184_169435335 | 0.11 |
XXyac-YX65C7_A.2 |
|
178090 |
0.03 |
| chr15_90380482_90380633 | 0.11 |
ENSG00000264966 |
. |
13396 |
0.11 |
| chr7_6433629_6433849 | 0.11 |
RAC1 |
ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
19569 |
0.16 |
| chr22_35615345_35615496 | 0.11 |
ENSG00000238584 |
. |
30345 |
0.16 |
| chr3_111527033_111527184 | 0.11 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
50919 |
0.15 |
| chr7_101584838_101585234 | 0.11 |
CTB-181H17.1 |
|
18360 |
0.23 |
| chr3_111457251_111457402 | 0.11 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
5982 |
0.25 |
| chr8_19041447_19041598 | 0.10 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
99282 |
0.08 |
| chr16_68694457_68694608 | 0.10 |
RP11-615I2.2 |
|
14461 |
0.13 |
| chr4_5640825_5640976 | 0.10 |
EVC2 |
Ellis van Creveld syndrome 2 |
69394 |
0.11 |
| chr5_73926315_73926659 | 0.10 |
HEXB |
hexosaminidase B (beta polypeptide) |
9361 |
0.21 |
| chr5_114750713_114750864 | 0.10 |
CCDC112 |
coiled-coil domain containing 112 |
118260 |
0.05 |
| chr15_67343283_67343434 | 0.10 |
SMAD3 |
SMAD family member 3 |
12743 |
0.28 |
| chr3_189840089_189840240 | 0.10 |
LEPREL1 |
leprecan-like 1 |
62 |
0.96 |
| chr5_72618007_72618158 | 0.10 |
FOXD1 |
forkhead box D1 |
126270 |
0.05 |
| chr6_86128809_86128960 | 0.10 |
NT5E |
5'-nucleotidase, ecto (CD73) |
30925 |
0.23 |
| chr3_167659356_167659607 | 0.10 |
SERPINI1 |
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
134455 |
0.05 |
| chr4_5384180_5384331 | 0.10 |
ENSG00000252266 |
. |
44456 |
0.18 |
| chr1_156877458_156877656 | 0.10 |
LRRC71 |
leucine rich repeat containing 71 |
12885 |
0.12 |
| chr21_45029813_45029964 | 0.10 |
HSF2BP |
heat shock transcription factor 2 binding protein |
48137 |
0.14 |
| chr2_3583541_3583693 | 0.10 |
RP13-512J5.1 |
Uncharacterized protein |
11930 |
0.12 |
| chr8_32573551_32573702 | 0.10 |
NRG1 |
neuregulin 1 |
5645 |
0.34 |
| chr2_99422286_99422437 | 0.10 |
ENSG00000201070 |
. |
23488 |
0.18 |
| chr1_27627251_27627402 | 0.10 |
TMEM222 |
transmembrane protein 222 |
21325 |
0.11 |
| chr3_100762310_100762461 | 0.10 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
50026 |
0.16 |
| chr8_92330361_92330512 | 0.10 |
SLC26A7 |
solute carrier family 26 (anion exchanger), member 7 |
22584 |
0.24 |
| chr15_60671550_60671832 | 0.10 |
ANXA2 |
annexin A2 |
4987 |
0.3 |
| chr9_95267331_95267483 | 0.10 |
ASPN |
asporin |
22619 |
0.15 |
| chr2_85817841_85818253 | 0.10 |
RNF181 |
ring finger protein 181 |
4801 |
0.1 |
| chr6_117823837_117823988 | 0.10 |
DCBLD1 |
discoidin, CUB and LCCL domain containing 1 |
20087 |
0.17 |
| chr8_103550021_103550172 | 0.10 |
ENSG00000252593 |
. |
4529 |
0.2 |
| chr14_85883772_85884074 | 0.10 |
RP11-497E19.2 |
Uncharacterized protein |
111020 |
0.07 |
| chr2_238106047_238106302 | 0.10 |
AC112715.2 |
Uncharacterized protein |
59560 |
0.13 |
| chr9_14159253_14159404 | 0.10 |
NFIB |
nuclear factor I/B |
21463 |
0.29 |
| chr4_146800249_146800400 | 0.10 |
RP11-181K12.2 |
|
46054 |
0.15 |
| chr15_36893589_36893740 | 0.10 |
C15orf41 |
chromosome 15 open reading frame 41 |
6563 |
0.31 |
| chr6_157950388_157950617 | 0.10 |
ENSG00000266617 |
. |
338 |
0.93 |
| chr14_73705630_73705816 | 0.10 |
PAPLN |
papilin, proteoglycan-like sulfated glycoprotein |
601 |
0.67 |
| chr20_19954474_19955311 | 0.10 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
42868 |
0.15 |
| chr2_101403389_101403540 | 0.10 |
NPAS2 |
neuronal PAS domain protein 2 |
33150 |
0.18 |
| chr2_33355690_33355841 | 0.10 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
3708 |
0.35 |
| chr15_36721035_36721186 | 0.10 |
C15orf41 |
chromosome 15 open reading frame 41 |
150702 |
0.05 |
| chr8_19040917_19041068 | 0.10 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
98752 |
0.08 |
| chr1_116213540_116213691 | 0.10 |
VANGL1 |
VANGL planar cell polarity protein 1 |
19592 |
0.2 |
| chr15_31898048_31898199 | 0.10 |
OTUD7A |
OTU domain containing 7A |
49419 |
0.19 |
| chr11_119331156_119331307 | 0.10 |
THY1 |
Thy-1 cell surface antigen |
35536 |
0.11 |
| chr12_95196881_95197032 | 0.10 |
ENSG00000208038 |
. |
31218 |
0.23 |
| chr14_103480838_103481545 | 0.10 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
42608 |
0.12 |
| chr15_39575982_39576133 | 0.10 |
RP11-624L4.1 |
|
9973 |
0.26 |
| chr3_105077778_105077929 | 0.10 |
ALCAM |
activated leukocyte cell adhesion molecule |
7900 |
0.34 |
| chr12_66002465_66002616 | 0.10 |
HMGA2 |
high mobility group AT-hook 2 |
215371 |
0.02 |
| chr18_47257293_47257444 | 0.10 |
ACAA2 |
acetyl-CoA acyltransferase 2 |
80533 |
0.07 |
| chr16_89947688_89947839 | 0.10 |
TCF25 |
transcription factor 25 (basic helix-loop-helix) |
2039 |
0.23 |
| chr12_65918531_65918682 | 0.10 |
MSRB3 |
methionine sulfoxide reductase B3 |
197951 |
0.03 |
| chr1_157998665_157998816 | 0.10 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
3400 |
0.28 |
| chr16_84133396_84133731 | 0.10 |
MBTPS1 |
membrane-bound transcription factor peptidase, site 1 |
16837 |
0.12 |
| chr11_129509524_129509675 | 0.10 |
TMEM45B |
transmembrane protein 45B |
176115 |
0.03 |
| chr11_101991203_101991354 | 0.10 |
YAP1 |
Yes-associated protein 1 |
8033 |
0.18 |
| chr13_30082723_30082874 | 0.10 |
MTUS2-AS1 |
MTUS2 antisense RNA 1 |
18556 |
0.25 |
| chr15_70087017_70087168 | 0.10 |
ENSG00000215958 |
. |
9249 |
0.28 |
| chr6_36819266_36819870 | 0.10 |
CPNE5 |
copine V |
11790 |
0.17 |
| chr9_89891782_89892083 | 0.10 |
ENSG00000212421 |
. |
16567 |
0.28 |
| chr18_43344332_43344486 | 0.10 |
RP11-116O18.3 |
|
17229 |
0.18 |
| chr7_70175760_70175911 | 0.10 |
AUTS2 |
autism susceptibility candidate 2 |
18290 |
0.31 |
| chr4_123703753_123703904 | 0.10 |
FGF2 |
fibroblast growth factor 2 (basic) |
44035 |
0.13 |
| chr20_33301699_33301850 | 0.10 |
TP53INP2 |
tumor protein p53 inducible nuclear protein 2 |
9235 |
0.21 |
| chr18_11501149_11501300 | 0.10 |
NPIPB1P |
nuclear pore complex interacting protein family, member B1, pseudogene |
138236 |
0.05 |
| chr8_19173658_19173809 | 0.10 |
SH2D4A |
SH2 domain containing 4A |
2246 |
0.46 |
| chr12_109044211_109044362 | 0.10 |
CORO1C |
coronin, actin binding protein, 1C |
1835 |
0.26 |
| chr16_58597342_58597493 | 0.10 |
ENSG00000206952 |
. |
3582 |
0.17 |
| chr20_33514132_33514283 | 0.10 |
GSS |
glutathione synthetase |
20325 |
0.15 |
| chr15_71587319_71587470 | 0.10 |
RP11-592N21.2 |
|
47269 |
0.17 |
| chr14_72164592_72164743 | 0.10 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
79194 |
0.11 |
| chr6_82724453_82724882 | 0.10 |
ENSG00000223044 |
. |
195490 |
0.03 |
| chr8_98944366_98944517 | 0.10 |
MATN2 |
matrilin 2 |
788 |
0.7 |
| chr1_48960343_48960494 | 0.09 |
SPATA6 |
spermatogenesis associated 6 |
22573 |
0.23 |
| chr1_201199232_201199383 | 0.09 |
RP11-567E21.3 |
|
7056 |
0.18 |
| chr18_8982789_8982940 | 0.09 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
119764 |
0.05 |
| chr3_73629408_73629559 | 0.09 |
PDZRN3 |
PDZ domain containing ring finger 3 |
10871 |
0.21 |
| chr6_52380007_52380403 | 0.09 |
TRAM2 |
translocation associated membrane protein 2 |
61508 |
0.12 |
| chr4_7137177_7137328 | 0.09 |
ENSG00000200867 |
. |
23126 |
0.16 |
| chr14_62013906_62014133 | 0.09 |
RP11-47I22.1 |
|
2014 |
0.33 |
| chr7_135446779_135447012 | 0.09 |
FAM180A |
family with sequence similarity 180, member A |
13301 |
0.17 |
| chr2_173792428_173792728 | 0.09 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
431 |
0.9 |
| chr8_117334188_117334602 | 0.09 |
ENSG00000264815 |
. |
34807 |
0.24 |
| chr3_137844512_137844702 | 0.09 |
A4GNT |
alpha-1,4-N-acetylglucosaminyltransferase |
6622 |
0.2 |
| chr3_99569503_99569951 | 0.09 |
FILIP1L |
filamin A interacting protein 1-like |
192 |
0.96 |
| chr3_69439182_69439463 | 0.09 |
FRMD4B |
FERM domain containing 4B |
3892 |
0.36 |
| chr17_29395281_29395436 | 0.09 |
RP11-271K11.5 |
|
17314 |
0.11 |
| chr12_110707275_110707426 | 0.09 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
11211 |
0.23 |
| chr8_29594762_29594913 | 0.09 |
ENSG00000221003 |
. |
191284 |
0.03 |
| chr6_155545820_155546032 | 0.09 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
7826 |
0.22 |
| chr6_45576539_45576690 | 0.09 |
ENSG00000252738 |
. |
37227 |
0.22 |
| chr11_94525821_94526186 | 0.09 |
AMOTL1 |
angiomotin like 1 |
24466 |
0.21 |
| chr9_94181980_94182457 | 0.09 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
3926 |
0.34 |
| chr7_36343272_36343423 | 0.09 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
6594 |
0.18 |
| chr20_19983080_19983231 | 0.09 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
14605 |
0.2 |
| chr6_24428860_24429018 | 0.09 |
MRS2 |
MRS2 magnesium transporter |
25519 |
0.16 |
| chr8_118923064_118923215 | 0.09 |
EXT1 |
exostosin glycosyltransferase 1 |
199514 |
0.03 |
| chr10_13908472_13908623 | 0.09 |
FRMD4A |
FERM domain containing 4A |
7635 |
0.26 |
| chr8_95323156_95323307 | 0.09 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
48653 |
0.15 |
| chr1_183028779_183028930 | 0.09 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
36259 |
0.16 |
| chr6_125325392_125325543 | 0.09 |
RNF217 |
ring finger protein 217 |
20953 |
0.23 |
| chr4_20466292_20466443 | 0.09 |
ENSG00000207732 |
. |
63531 |
0.12 |
| chr10_48276265_48276416 | 0.09 |
ANXA8 |
annexin A8 |
20987 |
0.13 |
| chr6_35463302_35463453 | 0.09 |
TEAD3 |
TEA domain family member 3 |
1350 |
0.4 |
| chr17_45392303_45392454 | 0.09 |
EFCAB13 |
EF-hand calcium binding domain 13 |
8278 |
0.16 |
| chr1_183030232_183030383 | 0.09 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
35372 |
0.16 |
| chr14_105558499_105558650 | 0.09 |
GPR132 |
G protein-coupled receptor 132 |
26792 |
0.16 |
| chr9_119312381_119312532 | 0.09 |
RP11-67K19.3 |
|
18272 |
0.22 |
| chr15_59462828_59463043 | 0.09 |
ENSG00000253030 |
. |
526 |
0.66 |
| chr1_23729323_23729474 | 0.09 |
TCEA3 |
transcription elongation factor A (SII), 3 |
15486 |
0.13 |
| chr1_20390393_20390544 | 0.09 |
PLA2G5 |
phospholipase A2, group V |
6233 |
0.2 |
| chr15_47816021_47816320 | 0.09 |
RP11-142J21.2 |
|
1196 |
0.53 |
| chr12_57546173_57546324 | 0.09 |
RP11-545N8.3 |
|
4846 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.0 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |