Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
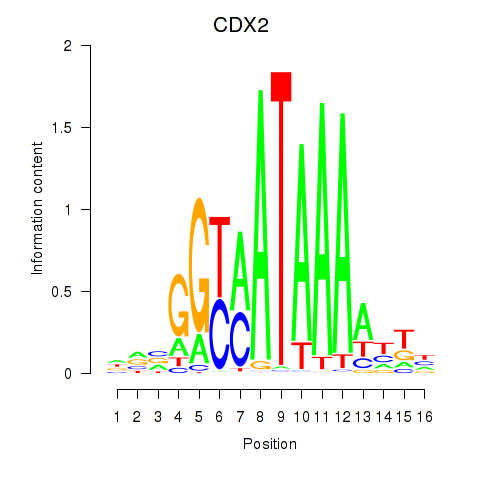
Results for CDX2
Z-value: 0.69
Transcription factors associated with CDX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX2
|
ENSG00000165556.9 | caudal type homeobox 2 |
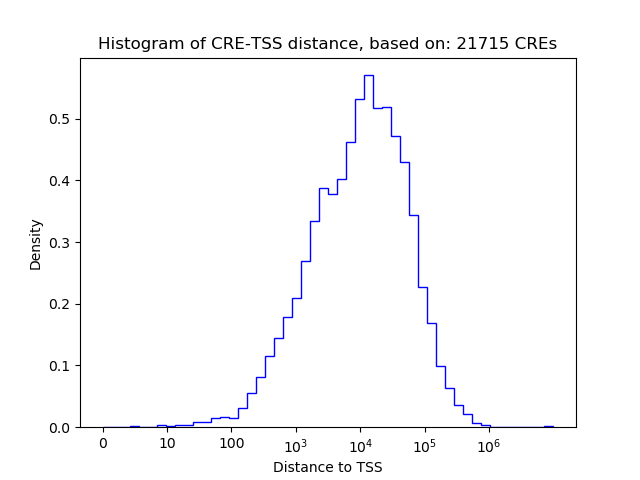
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_28553030_28553181 | CDX2 | 7829 | 0.144025 | 0.72 | 2.8e-02 | Click! |
| chr13_28542993_28543207 | CDX2 | 2176 | 0.236105 | 0.68 | 4.4e-02 | Click! |
| chr13_28542609_28542846 | CDX2 | 2549 | 0.211548 | 0.66 | 5.5e-02 | Click! |
| chr13_28534615_28535489 | CDX2 | 10224 | 0.136555 | 0.65 | 5.6e-02 | Click! |
| chr13_28535904_28536055 | CDX2 | 9297 | 0.138559 | 0.65 | 5.8e-02 | Click! |
Activity of the CDX2 motif across conditions
Conditions sorted by the z-value of the CDX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
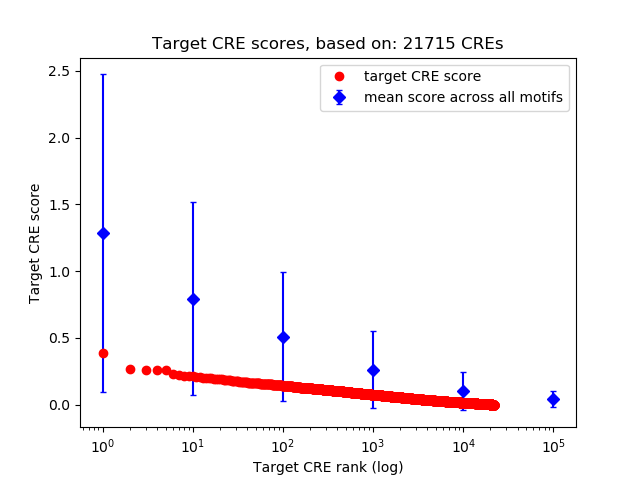
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_109582380_109582745 | 0.38 |
EDAR |
ectodysplasin A receptor |
23163 |
0.25 |
| chr1_81780447_81780600 | 0.27 |
LPHN2 |
latrophilin 2 |
8672 |
0.29 |
| chr1_101437966_101438267 | 0.26 |
SLC30A7 |
solute carrier family 30 (zinc transporter), member 7 |
10732 |
0.14 |
| chr8_66861197_66861965 | 0.26 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
72214 |
0.11 |
| chr5_122865285_122865436 | 0.26 |
CSNK1G3 |
casein kinase 1, gamma 3 |
15749 |
0.23 |
| chr6_12037649_12037832 | 0.23 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
21920 |
0.24 |
| chr1_160230794_160231302 | 0.22 |
RP11-574F21.2 |
|
486 |
0.45 |
| chr5_175974700_175974851 | 0.22 |
ENSG00000243959 |
. |
48 |
0.95 |
| chr2_48543908_48544266 | 0.22 |
FOXN2 |
forkhead box N2 |
2238 |
0.35 |
| chr12_111613238_111613505 | 0.22 |
CUX2 |
cut-like homeobox 2 |
76121 |
0.1 |
| chr3_69508196_69508347 | 0.21 |
FRMD4B |
FERM domain containing 4B |
72841 |
0.13 |
| chr4_143621409_143621560 | 0.21 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
139662 |
0.05 |
| chr10_82251750_82251998 | 0.20 |
TSPAN14 |
tetraspanin 14 |
32816 |
0.15 |
| chr1_6652266_6652423 | 0.20 |
KLHL21 |
kelch-like family member 21 |
7532 |
0.11 |
| chr6_11258753_11259646 | 0.20 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
26284 |
0.22 |
| chrX_64946026_64946177 | 0.20 |
MSN |
moesin |
58564 |
0.16 |
| chr8_66864395_66864582 | 0.20 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
69307 |
0.12 |
| chr8_96157942_96158334 | 0.19 |
PLEKHF2 |
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
11834 |
0.21 |
| chr4_175618125_175618276 | 0.19 |
GLRA3 |
glycine receptor, alpha 3 |
132178 |
0.05 |
| chr1_116673236_116673387 | 0.19 |
MAB21L3 |
mab-21-like 3 (C. elegans) |
18935 |
0.25 |
| chr18_3010170_3010321 | 0.19 |
LPIN2 |
lipin 2 |
1700 |
0.33 |
| chr2_9683437_9683847 | 0.19 |
ENSG00000238462 |
. |
11168 |
0.16 |
| chr4_141164391_141164692 | 0.18 |
SCOC |
short coiled-coil protein |
13899 |
0.21 |
| chr14_44060665_44060966 | 0.18 |
FSCB |
fibrous sheath CABYR binding protein |
915667 |
0.0 |
| chr12_93638767_93638982 | 0.18 |
ENSG00000238361 |
. |
20634 |
0.16 |
| chr12_11821489_11821646 | 0.18 |
ETV6 |
ets variant 6 |
18779 |
0.25 |
| chr10_89850893_89851084 | 0.18 |
ENSG00000200891 |
. |
96536 |
0.08 |
| chr14_38546372_38546618 | 0.18 |
CTD-2058B24.2 |
|
13868 |
0.25 |
| chr4_36243199_36243543 | 0.18 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
2190 |
0.3 |
| chr2_68404077_68404429 | 0.17 |
RP11-474G23.2 |
|
2712 |
0.22 |
| chr15_92455349_92455602 | 0.17 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
58124 |
0.15 |
| chr21_16056596_16056882 | 0.17 |
AF165138.7 |
Protein LOC388813 |
25597 |
0.23 |
| chr5_158526590_158526785 | 0.17 |
EBF1 |
early B-cell factor 1 |
14 |
0.99 |
| chr9_126664569_126664720 | 0.17 |
DENND1A |
DENN/MADD domain containing 1A |
27742 |
0.2 |
| chr1_14035183_14035334 | 0.17 |
PRDM2 |
PR domain containing 2, with ZNF domain |
3908 |
0.25 |
| chr2_120685704_120685855 | 0.17 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
1573 |
0.44 |
| chr1_100911531_100911742 | 0.17 |
RP5-837M10.4 |
|
39917 |
0.15 |
| chr2_145128325_145128476 | 0.17 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
38317 |
0.2 |
| chr2_48545707_48546468 | 0.17 |
FOXN2 |
forkhead box N2 |
4238 |
0.26 |
| chr10_126409005_126409284 | 0.17 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
16391 |
0.17 |
| chr1_108440044_108440852 | 0.17 |
VAV3-AS1 |
VAV3 antisense RNA 1 |
66617 |
0.11 |
| chr13_77318791_77318984 | 0.17 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
141638 |
0.05 |
| chr10_101942748_101942988 | 0.17 |
ERLIN1 |
ER lipid raft associated 1 |
2918 |
0.19 |
| chr2_12849849_12850137 | 0.17 |
TRIB2 |
tribbles pseudokinase 2 |
7022 |
0.27 |
| chr1_81673664_81673948 | 0.16 |
ENSG00000223026 |
. |
43668 |
0.2 |
| chr17_28658400_28658590 | 0.16 |
TMIGD1 |
transmembrane and immunoglobulin domain containing 1 |
2570 |
0.19 |
| chr7_127749161_127749327 | 0.16 |
ENSG00000207588 |
. |
27331 |
0.2 |
| chr15_31643081_31643271 | 0.16 |
KLF13 |
Kruppel-like factor 13 |
11040 |
0.3 |
| chr4_2487124_2487508 | 0.16 |
RNF4 |
ring finger protein 4 |
4222 |
0.21 |
| chr14_91479889_91480344 | 0.16 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
46362 |
0.13 |
| chr16_81530462_81530613 | 0.16 |
CMIP |
c-Maf inducing protein |
1583 |
0.51 |
| chr14_75735225_75735582 | 0.16 |
RP11-293M10.1 |
Uncharacterized protein |
583 |
0.61 |
| chr1_89751943_89752616 | 0.16 |
GBP5 |
guanylate binding protein 5 |
13735 |
0.19 |
| chr11_46366304_46366455 | 0.16 |
DGKZ |
diacylglycerol kinase, zeta |
438 |
0.79 |
| chr2_174763218_174763492 | 0.16 |
SP3 |
Sp3 transcription factor |
65592 |
0.15 |
| chr9_20256564_20257102 | 0.16 |
ENSG00000221744 |
. |
38169 |
0.2 |
| chr2_152144415_152144634 | 0.16 |
NMI |
N-myc (and STAT) interactor |
1884 |
0.36 |
| chr11_128532326_128532477 | 0.16 |
RP11-744N12.3 |
|
23922 |
0.16 |
| chr2_65286677_65286830 | 0.16 |
CEP68 |
centrosomal protein 68kDa |
3154 |
0.23 |
| chr5_86377835_86378358 | 0.16 |
ENSG00000251951 |
. |
20567 |
0.22 |
| chr2_172950128_172950279 | 0.16 |
DLX1 |
distal-less homeobox 1 |
61 |
0.98 |
| chr16_67398247_67398398 | 0.16 |
LRRC36 |
leucine rich repeat containing 36 |
17016 |
0.09 |
| chr1_204429917_204430068 | 0.16 |
PIK3C2B |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
6482 |
0.2 |
| chr14_23631200_23631351 | 0.16 |
ENSG00000202229 |
. |
5652 |
0.12 |
| chr16_55608208_55608359 | 0.16 |
CAPNS2 |
calpain, small subunit 2 |
7699 |
0.24 |
| chr19_36420808_36421170 | 0.15 |
LRFN3 |
leucine rich repeat and fibronectin type III domain containing 3 |
5463 |
0.1 |
| chr6_13724081_13724232 | 0.15 |
RANBP9 |
RAN binding protein 9 |
12360 |
0.2 |
| chr22_40877098_40877463 | 0.15 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
17842 |
0.16 |
| chr14_50547578_50547939 | 0.15 |
ENSG00000251792 |
. |
5942 |
0.16 |
| chr6_25432681_25432832 | 0.15 |
ENSG00000238322 |
. |
2752 |
0.28 |
| chr14_91837471_91837667 | 0.15 |
ENSG00000265856 |
. |
37512 |
0.16 |
| chr3_156534966_156535117 | 0.15 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
9048 |
0.29 |
| chr3_16984029_16984180 | 0.15 |
ENSG00000264818 |
. |
9416 |
0.23 |
| chr14_64226726_64226897 | 0.15 |
ENSG00000252749 |
. |
23649 |
0.17 |
| chr5_49735595_49735840 | 0.15 |
EMB |
embigin |
1467 |
0.6 |
| chr2_99312607_99313038 | 0.15 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
30337 |
0.18 |
| chr20_49132969_49133129 | 0.15 |
PTPN1 |
protein tyrosine phosphatase, non-receptor type 1 |
6129 |
0.18 |
| chr11_95940500_95940805 | 0.15 |
ENSG00000266192 |
. |
133950 |
0.05 |
| chr3_183893854_183894036 | 0.15 |
AP2M1 |
adaptor-related protein complex 2, mu 1 subunit |
621 |
0.57 |
| chr10_129856180_129856415 | 0.15 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
10463 |
0.28 |
| chr11_60047431_60047582 | 0.15 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
508 |
0.79 |
| chr13_74576794_74576945 | 0.15 |
KLF12 |
Kruppel-like factor 12 |
7683 |
0.34 |
| chr12_833209_833360 | 0.15 |
WNK1 |
WNK lysine deficient protein kinase 1 |
28475 |
0.16 |
| chr3_46973999_46974150 | 0.15 |
PTH1R |
parathyroid hormone 1 receptor |
29985 |
0.12 |
| chr3_171457874_171458025 | 0.15 |
PLD1 |
phospholipase D1, phosphatidylcholine-specific |
31200 |
0.18 |
| chr20_58714259_58714410 | 0.15 |
C20orf197 |
chromosome 20 open reading frame 197 |
83354 |
0.1 |
| chr12_96426880_96427227 | 0.15 |
LTA4H |
leukotriene A4 hydrolase |
2386 |
0.24 |
| chr6_4905786_4905958 | 0.15 |
CDYL |
chromodomain protein, Y-like |
15646 |
0.26 |
| chr17_76168269_76168420 | 0.15 |
SYNGR2 |
synaptogyrin 2 |
3105 |
0.14 |
| chr2_44408551_44408985 | 0.15 |
PPM1B |
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
12719 |
0.17 |
| chr1_92057392_92057549 | 0.15 |
CDC7 |
cell division cycle 7 |
90775 |
0.09 |
| chr13_107145729_107145927 | 0.15 |
EFNB2 |
ephrin-B2 |
41634 |
0.2 |
| chr15_67535265_67535775 | 0.15 |
AAGAB |
alpha- and gamma-adaptin binding protein |
311 |
0.91 |
| chr1_198581769_198581966 | 0.15 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
25934 |
0.21 |
| chr1_117359843_117360063 | 0.15 |
CD2 |
CD2 molecule |
62864 |
0.1 |
| chr19_35702667_35702989 | 0.15 |
FAM187B |
family with sequence similarity 187, member B |
16804 |
0.09 |
| chr19_42278293_42278624 | 0.14 |
AC011513.4 |
|
12343 |
0.12 |
| chr2_86413845_86413996 | 0.14 |
ENSG00000265420 |
. |
6311 |
0.13 |
| chr11_46535797_46536138 | 0.14 |
ENSG00000265014 |
. |
62528 |
0.09 |
| chr9_8622087_8622238 | 0.14 |
RP11-134K1.3 |
|
79390 |
0.11 |
| chr3_151974069_151974416 | 0.14 |
MBNL1 |
muscleblind-like splicing regulator 1 |
11587 |
0.22 |
| chr7_105713662_105713813 | 0.14 |
SYPL1 |
synaptophysin-like 1 |
24571 |
0.22 |
| chr12_54422690_54422841 | 0.14 |
HOXC6 |
homeobox C6 |
623 |
0.45 |
| chr7_7689968_7690119 | 0.14 |
RPA3 |
replication protein A3, 14kDa |
9440 |
0.26 |
| chr11_13482552_13482719 | 0.14 |
BTBD10 |
BTB (POZ) domain containing 10 |
2093 |
0.38 |
| chr7_75057844_75058027 | 0.14 |
POM121C |
POM121 transmembrane nucleoporin C |
12044 |
0.11 |
| chr15_60085307_60085458 | 0.14 |
BNIP2 |
BCL2/adenovirus E1B 19kDa interacting protein 2 |
103649 |
0.06 |
| chr13_48758888_48759039 | 0.14 |
ITM2B |
integral membrane protein 2B |
48331 |
0.15 |
| chr22_47176191_47176342 | 0.14 |
TBC1D22A |
TBC1 domain family, member 22A |
6442 |
0.21 |
| chr12_31903224_31903375 | 0.14 |
AMN1 |
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
21191 |
0.15 |
| chr4_16124268_16124434 | 0.14 |
PROM1 |
prominin 1 |
38350 |
0.16 |
| chr3_3149713_3149864 | 0.14 |
IL5RA |
interleukin 5 receptor, alpha |
1876 |
0.29 |
| chr3_121715323_121715474 | 0.14 |
ILDR1 |
immunoglobulin-like domain containing receptor 1 |
10750 |
0.19 |
| chr9_71459553_71459704 | 0.14 |
RP11-203L2.4 |
|
1437 |
0.5 |
| chr4_101351551_101351881 | 0.14 |
EMCN |
endomucin |
44518 |
0.2 |
| chr16_53495579_53495893 | 0.14 |
RBL2 |
retinoblastoma-like 2 (p130) |
11748 |
0.16 |
| chr1_167198816_167199102 | 0.14 |
POU2F1 |
POU class 2 homeobox 1 |
8816 |
0.19 |
| chr18_72344613_72344764 | 0.14 |
ZNF407 |
zinc finger protein 407 |
1712 |
0.5 |
| chr7_72705268_72705440 | 0.14 |
GTF2IRD2P1 |
GTF2I repeat domain containing 2 pseudogene 1 |
11211 |
0.14 |
| chr14_71466068_71466359 | 0.14 |
PCNX |
pecanex homolog (Drosophila) |
13535 |
0.29 |
| chr10_6644457_6644608 | 0.14 |
PRKCQ |
protein kinase C, theta |
22269 |
0.28 |
| chr1_38657052_38657335 | 0.14 |
ENSG00000265596 |
. |
102290 |
0.06 |
| chr1_202760210_202760361 | 0.14 |
KDM5B |
lysine (K)-specific demethylase 5B |
16142 |
0.13 |
| chr13_52390546_52391185 | 0.14 |
RP11-327P2.5 |
|
12432 |
0.19 |
| chr12_104865091_104865422 | 0.14 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
14477 |
0.26 |
| chr3_186763060_186763211 | 0.14 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
1604 |
0.46 |
| chr6_152269471_152269669 | 0.14 |
ESR1 |
estrogen receptor 1 |
67778 |
0.13 |
| chr11_116870995_116871424 | 0.14 |
ENSG00000264344 |
. |
15036 |
0.16 |
| chr2_161946247_161946703 | 0.14 |
TANK |
TRAF family member-associated NFKB activator |
46944 |
0.17 |
| chr7_20826220_20826440 | 0.14 |
SP8 |
Sp8 transcription factor |
175 |
0.97 |
| chr16_27650621_27650772 | 0.14 |
AC002551.1 |
|
3824 |
0.23 |
| chr2_213832657_213832808 | 0.14 |
ENSG00000266354 |
. |
41751 |
0.18 |
| chr7_37431624_37431775 | 0.13 |
ENSG00000200113 |
. |
27532 |
0.16 |
| chr20_1475362_1475513 | 0.13 |
SIRPB2 |
signal-regulatory protein beta 2 |
3310 |
0.17 |
| chr18_56355165_56355674 | 0.13 |
RP11-126O1.4 |
|
10573 |
0.15 |
| chr1_212111630_212111803 | 0.13 |
ENSG00000212187 |
. |
87187 |
0.07 |
| chr14_103908821_103908972 | 0.13 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
23155 |
0.14 |
| chr21_19153681_19154154 | 0.13 |
AL109761.5 |
|
11888 |
0.23 |
| chr6_106969864_106970230 | 0.13 |
AIM1 |
absent in melanoma 1 |
10317 |
0.2 |
| chr5_39185830_39185981 | 0.13 |
FYB |
FYN binding protein |
17224 |
0.25 |
| chr2_70779527_70779756 | 0.13 |
TGFA |
transforming growth factor, alpha |
981 |
0.62 |
| chr2_219031375_219031607 | 0.13 |
CXCR1 |
chemokine (C-X-C motif) receptor 1 |
227 |
0.91 |
| chr5_75565842_75566100 | 0.13 |
RP11-466P24.6 |
|
41316 |
0.2 |
| chr6_7439816_7439967 | 0.13 |
CAGE1 |
cancer antigen 1 |
49915 |
0.13 |
| chr11_77182415_77182566 | 0.13 |
DKFZP434E1119 |
|
1926 |
0.31 |
| chr5_179513527_179513811 | 0.13 |
RNF130 |
ring finger protein 130 |
14551 |
0.2 |
| chr15_52060726_52060931 | 0.13 |
CTD-2308G16.1 |
|
16146 |
0.13 |
| chr2_197022675_197023058 | 0.13 |
STK17B |
serine/threonine kinase 17b |
1495 |
0.42 |
| chr3_20093122_20093273 | 0.13 |
KAT2B |
K(lysine) acetyltransferase 2B |
11682 |
0.19 |
| chr13_99872623_99872781 | 0.13 |
ENSG00000201793 |
. |
14740 |
0.17 |
| chr8_38226618_38226769 | 0.13 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
11646 |
0.12 |
| chr13_27975238_27975401 | 0.13 |
ENSG00000201242 |
. |
1405 |
0.39 |
| chr14_99730566_99730874 | 0.13 |
AL109767.1 |
|
1435 |
0.45 |
| chr11_36402058_36402209 | 0.13 |
PRR5L |
proline rich 5 like |
3061 |
0.26 |
| chr5_98261626_98261777 | 0.13 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
539 |
0.8 |
| chr11_36429123_36429289 | 0.13 |
PRR5L |
proline rich 5 like |
6591 |
0.2 |
| chr5_94891308_94891836 | 0.13 |
ARSK |
arylsulfatase family, member K |
701 |
0.47 |
| chr5_108991988_108992139 | 0.13 |
ENSG00000266090 |
. |
29218 |
0.18 |
| chr5_167570316_167570766 | 0.13 |
CTB-178M22.1 |
|
44183 |
0.16 |
| chr9_134532062_134532585 | 0.13 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
30988 |
0.18 |
| chr6_97497693_97497949 | 0.13 |
KLHL32 |
kelch-like family member 32 |
14683 |
0.28 |
| chr6_126154204_126154432 | 0.13 |
NCOA7-AS1 |
NCOA7 antisense RNA 1 |
14314 |
0.18 |
| chr17_48530281_48530435 | 0.13 |
CHAD |
chondroadherin |
12428 |
0.1 |
| chr8_41853450_41853601 | 0.13 |
KAT6A |
K(lysine) acetyltransferase 6A |
37644 |
0.15 |
| chr7_17973312_17973463 | 0.13 |
SNX13 |
sorting nexin 13 |
6704 |
0.28 |
| chr22_40783696_40783896 | 0.13 |
RP5-1042K10.10 |
|
425 |
0.81 |
| chr1_88364293_88364444 | 0.13 |
ENSG00000199318 |
. |
445312 |
0.01 |
| chr13_99566346_99566616 | 0.13 |
DOCK9 |
dedicator of cytokinesis 9 |
58243 |
0.13 |
| chr13_99723069_99723220 | 0.13 |
DOCK9 |
dedicator of cytokinesis 9 |
15516 |
0.19 |
| chr3_112692893_112693044 | 0.13 |
CD200R1 |
CD200 receptor 1 |
791 |
0.59 |
| chr17_58472537_58472688 | 0.13 |
C17orf64 |
chromosome 17 open reading frame 64 |
2822 |
0.26 |
| chr12_82336263_82336414 | 0.13 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
183006 |
0.03 |
| chr3_186729650_186729921 | 0.13 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
9858 |
0.24 |
| chr15_85934213_85934364 | 0.13 |
ENSG00000251891 |
. |
1173 |
0.52 |
| chr2_198007042_198007193 | 0.13 |
ANKRD44 |
ankyrin repeat domain 44 |
5511 |
0.27 |
| chr9_93570502_93570735 | 0.13 |
SYK |
spleen tyrosine kinase |
6409 |
0.34 |
| chr13_111225360_111225511 | 0.13 |
RAB20 |
RAB20, member RAS oncogene family |
11355 |
0.21 |
| chr6_42006686_42006952 | 0.13 |
CCND3 |
cyclin D3 |
9605 |
0.15 |
| chr9_123956488_123956761 | 0.13 |
RAB14 |
RAB14, member RAS oncogene family |
7523 |
0.16 |
| chr4_109453536_109453687 | 0.13 |
ENSG00000266046 |
. |
44685 |
0.17 |
| chr2_43373091_43373242 | 0.13 |
ENSG00000207087 |
. |
54534 |
0.14 |
| chr8_30501926_30502077 | 0.13 |
ENSG00000254172 |
. |
795 |
0.6 |
| chr12_71117144_71117295 | 0.12 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
31154 |
0.22 |
| chr8_41899359_41899510 | 0.12 |
ENSG00000238966 |
. |
1659 |
0.39 |
| chr10_35661911_35662062 | 0.12 |
CCNY |
cyclin Y |
36184 |
0.16 |
| chr14_68749923_68750260 | 0.12 |
ENSG00000243546 |
. |
46835 |
0.17 |
| chr1_175166360_175166539 | 0.12 |
KIAA0040 |
KIAA0040 |
4370 |
0.31 |
| chr18_25757142_25757385 | 0.12 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
147 |
0.98 |
| chr2_207854350_207854673 | 0.12 |
CPO |
carboxypeptidase O |
50233 |
0.15 |
| chr18_73122194_73122345 | 0.12 |
RP11-321M21.3 |
HCG1996301; Uncharacterized protein |
3302 |
0.31 |
| chr7_27252518_27252718 | 0.12 |
HOTTIP |
HOXA distal transcript antisense RNA |
12562 |
0.07 |
| chr22_42232265_42232534 | 0.12 |
RP5-821D11.7 |
|
1730 |
0.24 |
| chr4_166178779_166178930 | 0.12 |
ENSG00000200974 |
. |
5122 |
0.22 |
| chr10_8225843_8226238 | 0.12 |
GATA3 |
GATA binding protein 3 |
129271 |
0.06 |
| chr15_40410256_40410479 | 0.12 |
BMF |
Bcl2 modifying factor |
9274 |
0.16 |
| chr18_8787123_8787274 | 0.12 |
SOGA2 |
SOGA family member 2 |
2474 |
0.3 |
| chr1_111328214_111328365 | 0.12 |
ENSG00000199710 |
. |
21161 |
0.2 |
| chr3_56729846_56730094 | 0.12 |
FAM208A |
family with sequence similarity 208, member A |
12705 |
0.27 |
| chr2_241503394_241503545 | 0.12 |
RNPEPL1 |
arginyl aminopeptidase (aminopeptidase B)-like 1 |
1752 |
0.22 |
| chr2_201752130_201752281 | 0.12 |
PPIL3 |
peptidylprolyl isomerase (cyclophilin)-like 3 |
1097 |
0.27 |
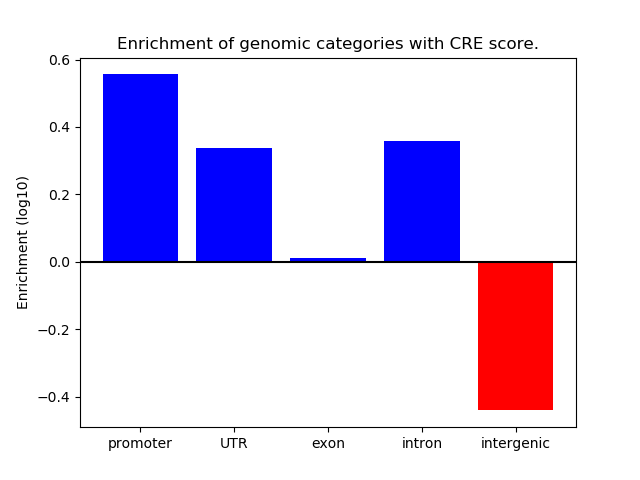
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.2 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.0 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.3 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.0 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:1901663 | quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0046348 | amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.0 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.0 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.0 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0060487 | lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.0 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0032069 | regulation of nuclease activity(GO:0032069) |
| 0.0 | 0.2 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0070076 | histone lysine demethylation(GO:0070076) |
| 0.0 | 0.1 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0050854 | regulation of antigen receptor-mediated signaling pathway(GO:0050854) |
| 0.0 | 0.0 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0046643 | regulation of gamma-delta T cell activation(GO:0046643) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.1 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.0 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.0 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.0 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.0 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.0 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.0 | REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | Genes involved in Class I MHC mediated antigen processing & presentation |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.1 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.1 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |