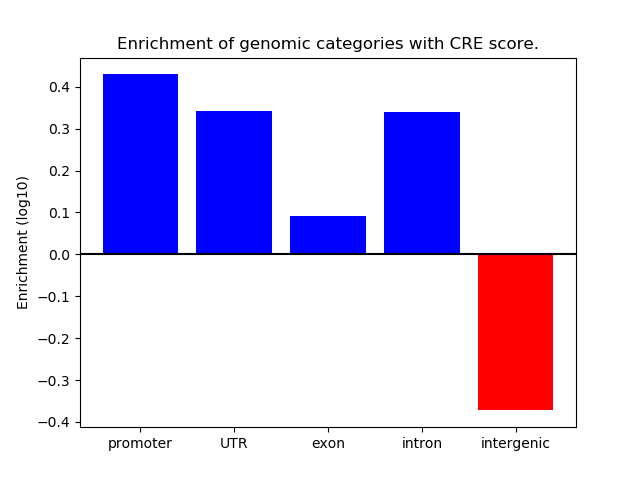
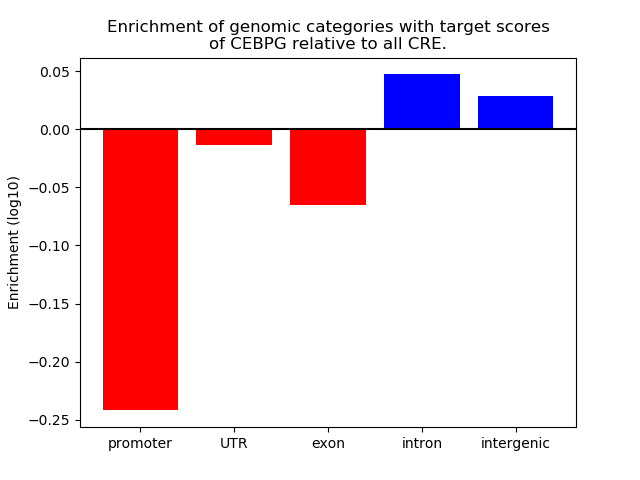
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
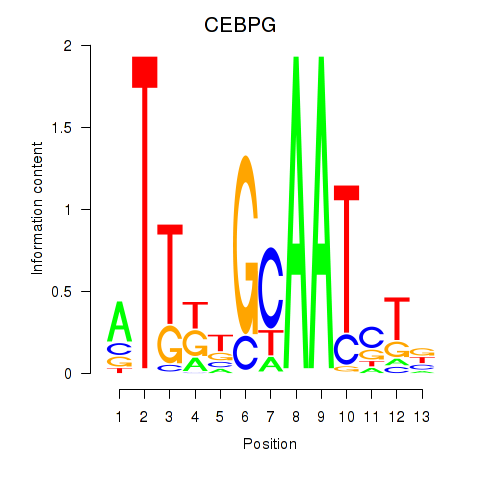
Results for CEBPG
Z-value: 0.59
Transcription factors associated with CEBPG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPG
|
ENSG00000153879.4 | CCAAT enhancer binding protein gamma |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_33859090_33859241 | CEBPG | 5071 | 0.212328 | -0.69 | 4.0e-02 | Click! |
| chr19_33886220_33886371 | CEBPG | 21077 | 0.186981 | -0.67 | 4.8e-02 | Click! |
| chr19_33859334_33859485 | CEBPG | 4827 | 0.215335 | -0.56 | 1.2e-01 | Click! |
| chr19_33888075_33888226 | CEBPG | 22932 | 0.183493 | -0.35 | 3.5e-01 | Click! |
| chr19_33887099_33887250 | CEBPG | 21956 | 0.185299 | -0.33 | 3.8e-01 | Click! |
Activity of the CEBPG motif across conditions
Conditions sorted by the z-value of the CEBPG motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
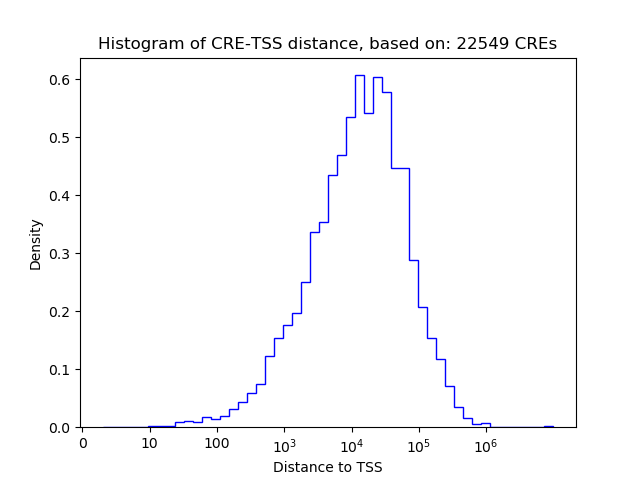
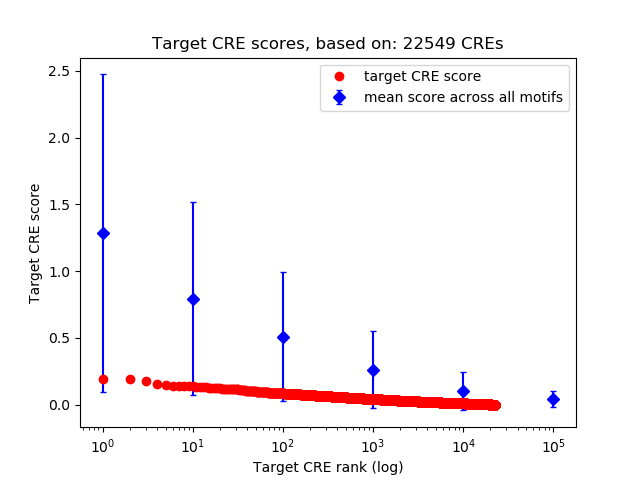
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_158899724_158899977 | 0.19 |
UPP2 |
uridine phosphorylase 2 |
48139 |
0.14 |
| chr6_147090769_147091051 | 0.19 |
ADGB |
androglobin |
665 |
0.81 |
| chr2_231845610_231845781 | 0.18 |
SPATA3 |
spermatogenesis associated 3 |
15141 |
0.15 |
| chr2_201958452_201958603 | 0.16 |
ENSG00000252148 |
. |
13962 |
0.12 |
| chr9_111604840_111604991 | 0.15 |
ACTL7B |
actin-like 7B |
14324 |
0.2 |
| chr12_65045752_65045922 | 0.14 |
RP11-338E21.3 |
|
2962 |
0.18 |
| chr20_5668640_5669158 | 0.14 |
C20orf196 |
chromosome 20 open reading frame 196 |
62140 |
0.12 |
| chr4_154627589_154627740 | 0.14 |
TLR2 |
toll-like receptor 2 |
5012 |
0.2 |
| chr21_45509017_45509467 | 0.14 |
PWP2 |
PWP2 periodic tryptophan protein homolog (yeast) |
17929 |
0.16 |
| chr2_182256895_182257046 | 0.14 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
64964 |
0.13 |
| chrX_53962631_53962782 | 0.14 |
ENSG00000201618 |
. |
27218 |
0.22 |
| chr7_155446676_155446874 | 0.14 |
RBM33 |
RNA binding motif protein 33 |
9379 |
0.22 |
| chr1_169575791_169576071 | 0.13 |
SELP |
selectin P (granule membrane protein 140kDa, antigen CD62) |
12526 |
0.2 |
| chr2_68639578_68639939 | 0.13 |
AC015969.3 |
|
47042 |
0.11 |
| chr12_8229057_8229208 | 0.13 |
NECAP1 |
NECAP endocytosis associated 1 |
5675 |
0.16 |
| chr15_91799071_91799222 | 0.13 |
SV2B |
synaptic vesicle glycoprotein 2B |
30046 |
0.23 |
| chr4_84032777_84033059 | 0.12 |
PLAC8 |
placenta-specific 8 |
1922 |
0.4 |
| chr2_68597035_68597533 | 0.12 |
AC015969.3 |
|
4568 |
0.19 |
| chr12_14587365_14587516 | 0.12 |
ATF7IP |
activating transcription factor 7 interacting protein |
10745 |
0.24 |
| chr16_23879059_23879244 | 0.12 |
PRKCB |
protein kinase C, beta |
30607 |
0.2 |
| chr9_7977391_7977713 | 0.12 |
TMEM261 |
transmembrane protein 261 |
177485 |
0.04 |
| chr4_7648871_7649036 | 0.12 |
SORCS2 |
sortilin-related VPS10 domain containing receptor 2 |
87054 |
0.09 |
| chr4_87915586_87915883 | 0.12 |
AFF1 |
AF4/FMR2 family, member 1 |
12406 |
0.26 |
| chr12_68751196_68751376 | 0.12 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
25125 |
0.24 |
| chr4_13617345_13617663 | 0.12 |
BOD1L1 |
biorientation of chromosomes in cell division 1-like 1 |
11843 |
0.2 |
| chr21_16899077_16899228 | 0.12 |
ENSG00000212564 |
. |
87450 |
0.1 |
| chr18_66388930_66389081 | 0.12 |
TMX3 |
thioredoxin-related transmembrane protein 3 |
6470 |
0.21 |
| chr10_48531683_48532056 | 0.12 |
GDF10 |
growth differentiation factor 10 |
92893 |
0.07 |
| chr15_79887658_79887809 | 0.12 |
KIAA1024 |
KIAA1024 |
162875 |
0.03 |
| chr5_148415129_148415422 | 0.12 |
SH3TC2 |
SH3 domain and tetratricopeptide repeats 2 |
5834 |
0.18 |
| chr8_74585562_74585746 | 0.12 |
STAU2 |
staufen double-stranded RNA binding protein 2 |
64908 |
0.13 |
| chr8_101589648_101589799 | 0.11 |
SNX31 |
sorting nexin 31 |
6665 |
0.18 |
| chr9_33715106_33715257 | 0.11 |
TRBV26OR9-2 |
T cell receptor beta variable 26/OR9-2 (pseudogene) |
19416 |
0.14 |
| chr4_55667592_55667743 | 0.11 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
143582 |
0.04 |
| chr13_46579512_46579663 | 0.11 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
35782 |
0.16 |
| chr3_186724517_186724802 | 0.11 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
14984 |
0.23 |
| chr5_40387880_40388031 | 0.11 |
ENSG00000265615 |
. |
67535 |
0.13 |
| chr18_37166921_37167072 | 0.11 |
ENSG00000266148 |
. |
89689 |
0.1 |
| chr11_3911438_3911625 | 0.11 |
STIM1 |
stromal interaction molecule 1 |
2214 |
0.22 |
| chr6_114105204_114105355 | 0.11 |
ENSG00000253091 |
. |
2612 |
0.32 |
| chrX_65219579_65219921 | 0.11 |
RP6-159A1.3 |
|
157 |
0.96 |
| chr2_13118238_13118456 | 0.10 |
ENSG00000264370 |
. |
240854 |
0.02 |
| chr2_59892760_59892911 | 0.10 |
ENSG00000200101 |
. |
18079 |
0.24 |
| chr9_92141364_92141857 | 0.10 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
28565 |
0.19 |
| chr16_72051137_72051288 | 0.10 |
DHODH |
dihydroorotate dehydrogenase (quinone) |
8512 |
0.15 |
| chr1_108397007_108397158 | 0.10 |
ENSG00000265536 |
. |
78204 |
0.1 |
| chr7_139561651_139561816 | 0.10 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
32623 |
0.21 |
| chr20_20632126_20632277 | 0.10 |
RALGAPA2 |
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
11657 |
0.29 |
| chr5_53736395_53736669 | 0.10 |
HSPB3 |
heat shock 27kDa protein 3 |
14913 |
0.27 |
| chr15_90721896_90722171 | 0.10 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
6119 |
0.14 |
| chr8_19744288_19744439 | 0.10 |
LPL |
lipoprotein lipase |
14865 |
0.25 |
| chr5_50030005_50030180 | 0.10 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
66701 |
0.15 |
| chr2_135070977_135071198 | 0.10 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
59257 |
0.15 |
| chr2_7609287_7609438 | 0.10 |
ENSG00000221255 |
. |
107610 |
0.08 |
| chr10_115258928_115259188 | 0.10 |
HABP2 |
hyaluronan binding protein 2 |
51538 |
0.16 |
| chr11_104971720_104972111 | 0.10 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
243 |
0.33 |
| chr10_81933200_81933492 | 0.09 |
ANXA11 |
annexin A11 |
569 |
0.78 |
| chr15_86243186_86243337 | 0.09 |
RP11-815J21.1 |
|
1079 |
0.38 |
| chr3_118604005_118604156 | 0.09 |
IGSF11-AS1 |
IGSF11 antisense RNA 1 |
57840 |
0.15 |
| chr3_31440978_31441129 | 0.09 |
ENSG00000238727 |
. |
3624 |
0.36 |
| chrX_118629740_118630258 | 0.09 |
SLC25A5 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
27636 |
0.12 |
| chr7_29506906_29507057 | 0.09 |
CHN2 |
chimerin 2 |
12505 |
0.26 |
| chr3_114835975_114836126 | 0.09 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
30068 |
0.26 |
| chr1_202806183_202806334 | 0.09 |
RP11-480I12.4 |
Putative inactive alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase-like protein LOC641515 |
13873 |
0.11 |
| chrX_37646415_37646566 | 0.09 |
CYBB |
cytochrome b-245, beta polypeptide |
7173 |
0.25 |
| chr17_68339296_68339447 | 0.09 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
173695 |
0.03 |
| chr2_144157442_144157593 | 0.09 |
AC096558.1 |
|
80833 |
0.11 |
| chr12_11821669_11822031 | 0.09 |
ETV6 |
ets variant 6 |
19062 |
0.25 |
| chr2_120096690_120096841 | 0.09 |
C2orf76 |
chromosome 2 open reading frame 76 |
27493 |
0.16 |
| chr13_109597145_109597296 | 0.09 |
MYO16 |
myosin XVI |
58703 |
0.16 |
| chr1_229155807_229155958 | 0.09 |
RP5-1061H20.5 |
|
207427 |
0.02 |
| chr1_11815378_11815529 | 0.09 |
C1orf167 |
chromosome 1 open reading frame 167 |
6391 |
0.13 |
| chr2_135641742_135641893 | 0.09 |
ENSG00000263783 |
. |
26427 |
0.15 |
| chr7_114660124_114660310 | 0.09 |
MDFIC |
MyoD family inhibitor domain containing |
86293 |
0.1 |
| chr13_53712062_53712213 | 0.09 |
OLFM4 |
olfactomedin 4 |
109243 |
0.07 |
| chr15_48622427_48622580 | 0.09 |
DUT |
deoxyuridine triphosphatase |
705 |
0.58 |
| chr2_143908158_143909031 | 0.09 |
RP11-190J23.1 |
|
21147 |
0.22 |
| chr2_191018718_191018869 | 0.09 |
C2orf88 |
chromosome 2 open reading frame 88 |
3821 |
0.29 |
| chr7_135152018_135152169 | 0.09 |
CNOT4 |
CCR4-NOT transcription complex, subunit 4 |
29014 |
0.21 |
| chr2_44805499_44805827 | 0.09 |
ENSG00000252896 |
. |
203606 |
0.02 |
| chr13_32523758_32523999 | 0.09 |
EEF1DP3 |
eukaryotic translation elongation factor 1 delta pseudogene 3 |
2839 |
0.36 |
| chr5_173334399_173334550 | 0.09 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
14393 |
0.25 |
| chr2_197080013_197080164 | 0.09 |
ENSG00000239161 |
. |
42 |
0.98 |
| chr3_120311624_120311775 | 0.09 |
NDUFB4 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
3457 |
0.31 |
| chr1_172361702_172361853 | 0.09 |
DNM3 |
dynamin 3 |
13619 |
0.16 |
| chr6_36126532_36126683 | 0.09 |
ENSG00000221743 |
. |
18242 |
0.15 |
| chr13_47154815_47155055 | 0.09 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
27540 |
0.24 |
| chrX_147003132_147003283 | 0.09 |
FMR1 |
fragile X mental retardation 1 |
8514 |
0.15 |
| chr22_40842260_40842460 | 0.09 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
17062 |
0.15 |
| chr11_35325389_35325712 | 0.09 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
2475 |
0.3 |
| chr6_139446035_139446287 | 0.09 |
HECA |
headcase homolog (Drosophila) |
10088 |
0.26 |
| chr1_93028624_93028836 | 0.09 |
GFI1 |
growth factor independent 1 transcription repressor |
76297 |
0.1 |
| chr12_47604728_47604942 | 0.09 |
PCED1B |
PC-esterase domain containing 1B |
5217 |
0.24 |
| chr22_21198006_21198157 | 0.09 |
PI4KA |
phosphatidylinositol 4-kinase, catalytic, alpha |
14989 |
0.13 |
| chr16_53485634_53485785 | 0.09 |
RBL2 |
retinoblastoma-like 2 (p130) |
1721 |
0.3 |
| chr1_18395008_18395159 | 0.09 |
IGSF21 |
immunoglobin superfamily, member 21 |
39157 |
0.21 |
| chr2_99445353_99445801 | 0.09 |
ENSG00000238830 |
. |
11622 |
0.21 |
| chr3_193216992_193217143 | 0.09 |
ATP13A4-AS1 |
ATP13A4 antisense RNA 1 |
53935 |
0.12 |
| chr1_221335809_221335960 | 0.09 |
HLX |
H2.0-like homeobox |
281300 |
0.01 |
| chr2_41217129_41217280 | 0.08 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
379011 |
0.01 |
| chr7_32696237_32696388 | 0.08 |
ENSG00000207573 |
. |
76281 |
0.1 |
| chr11_44248403_44248554 | 0.08 |
ALX4 |
ALX homeobox 4 |
83238 |
0.09 |
| chr6_111692411_111692562 | 0.08 |
REV3L-IT1 |
REV3L intronic transcript 1 (non-protein coding) |
9676 |
0.21 |
| chr9_21252315_21252536 | 0.08 |
IFNA14 |
interferon, alpha 14 |
12447 |
0.12 |
| chr3_7794767_7794918 | 0.08 |
RP11-157E16.1 |
|
145258 |
0.05 |
| chr9_101513075_101513226 | 0.08 |
ANKS6 |
ankyrin repeat and sterile alpha motif domain containing 6 |
26558 |
0.18 |
| chr2_55091967_55092118 | 0.08 |
ENSG00000252507 |
. |
14616 |
0.25 |
| chr3_15331093_15331244 | 0.08 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
13616 |
0.14 |
| chr14_83934156_83934307 | 0.08 |
ENSG00000238561 |
. |
282497 |
0.01 |
| chr9_100866613_100866810 | 0.08 |
TRIM14 |
tripartite motif containing 14 |
11868 |
0.18 |
| chr12_32052086_32052476 | 0.08 |
ENSG00000252584 |
. |
16360 |
0.21 |
| chr4_123178885_123179036 | 0.08 |
KIAA1109 |
KIAA1109 |
2946 |
0.38 |
| chr3_152898855_152899006 | 0.08 |
ENSG00000265813 |
. |
18876 |
0.19 |
| chr8_101259438_101259589 | 0.08 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
55945 |
0.1 |
| chrX_30608424_30608610 | 0.08 |
CXorf21 |
chromosome X open reading frame 21 |
12556 |
0.23 |
| chr6_35987038_35987189 | 0.08 |
SLC26A8 |
solute carrier family 26 (anion exchanger), member 8 |
5157 |
0.21 |
| chr7_139060065_139060216 | 0.08 |
C7orf55-LUC7L2 |
C7orf55-LUC7L2 readthrough |
980 |
0.49 |
| chr9_123200418_123200569 | 0.08 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
9857 |
0.31 |
| chr6_137111692_137111843 | 0.08 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
1889 |
0.37 |
| chr17_74074475_74074626 | 0.08 |
ZACN |
zinc activated ligand-gated ion channel |
713 |
0.51 |
| chrX_15927183_15927334 | 0.08 |
ENSG00000200566 |
. |
7168 |
0.25 |
| chr20_32438754_32438905 | 0.08 |
CHMP4B |
charged multivesicular body protein 4B |
39719 |
0.13 |
| chr10_32248830_32248981 | 0.08 |
ARHGAP12 |
Rho GTPase activating protein 12 |
31163 |
0.17 |
| chr12_93528524_93528749 | 0.08 |
RP11-511B23.2 |
|
4231 |
0.26 |
| chr4_74625570_74625721 | 0.08 |
IL8 |
interleukin 8 |
19387 |
0.2 |
| chr10_82251255_82251532 | 0.08 |
TSPAN14 |
tetraspanin 14 |
32335 |
0.15 |
| chr5_14544889_14545095 | 0.08 |
FAM105A |
family with sequence similarity 105, member A |
36892 |
0.19 |
| chr18_77452829_77452980 | 0.08 |
RP11-567M16.5 |
|
10747 |
0.23 |
| chr9_78711137_78711288 | 0.08 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
319 |
0.94 |
| chr19_30748039_30748190 | 0.08 |
AC005597.1 |
|
28492 |
0.21 |
| chr2_43382252_43382437 | 0.08 |
ENSG00000207087 |
. |
63712 |
0.12 |
| chr3_113473365_113473635 | 0.08 |
ATP6V1A |
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
7603 |
0.19 |
| chr12_123304867_123305018 | 0.08 |
HIP1R |
huntingtin interacting protein 1 related |
15031 |
0.15 |
| chr1_36018860_36019118 | 0.08 |
KIAA0319L |
KIAA0319-like |
1599 |
0.31 |
| chr8_64093598_64093888 | 0.08 |
YTHDF3 |
YTH domain family, member 3 |
6345 |
0.27 |
| chr12_9191656_9191852 | 0.08 |
A2M-AS1 |
A2M antisense RNA 1 |
26019 |
0.14 |
| chr20_39667687_39667838 | 0.08 |
TOP1 |
topoisomerase (DNA) I |
10304 |
0.22 |
| chr5_73333262_73333413 | 0.08 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
151609 |
0.04 |
| chr10_112027697_112027848 | 0.08 |
SMNDC1 |
survival motor neuron domain containing 1 |
36683 |
0.16 |
| chr1_81679030_81679181 | 0.08 |
ENSG00000223026 |
. |
38369 |
0.22 |
| chr12_82336825_82337040 | 0.08 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
183600 |
0.03 |
| chr7_37009785_37010186 | 0.08 |
ELMO1 |
engulfment and cell motility 1 |
14680 |
0.2 |
| chr8_106142283_106142434 | 0.08 |
RP11-574O7.1 |
|
58476 |
0.16 |
| chr20_52357947_52358098 | 0.08 |
ENSG00000238468 |
. |
72725 |
0.11 |
| chr6_113807765_113807916 | 0.08 |
ENSG00000222677 |
. |
28349 |
0.25 |
| chr12_29543998_29544228 | 0.08 |
OVCH1-AS1 |
OVCH1 antisense RNA 1 |
1442 |
0.41 |
| chr7_37484804_37484955 | 0.08 |
ELMO1 |
engulfment and cell motility 1 |
3674 |
0.24 |
| chr5_6792995_6793146 | 0.08 |
ENSG00000200243 |
. |
55505 |
0.15 |
| chr4_41867501_41867667 | 0.08 |
TMEM33 |
transmembrane protein 33 |
69553 |
0.1 |
| chr10_71922318_71922469 | 0.08 |
SAR1A |
SAR1 homolog A (S. cerevisiae) |
394 |
0.85 |
| chr6_157144634_157144785 | 0.08 |
RP11-230C9.3 |
|
43120 |
0.15 |
| chr12_68761789_68762303 | 0.08 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
35885 |
0.21 |
| chr2_128927617_128927768 | 0.08 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
9668 |
0.24 |
| chr2_197070639_197070869 | 0.08 |
ENSG00000239161 |
. |
9292 |
0.19 |
| chr6_151120712_151120863 | 0.08 |
MTHFD1L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
65898 |
0.12 |
| chr2_64449988_64450270 | 0.08 |
AC074289.1 |
|
4866 |
0.3 |
| chr9_92147259_92147508 | 0.08 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
34338 |
0.17 |
| chrX_20229909_20230060 | 0.08 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
6995 |
0.22 |
| chr15_68422212_68422419 | 0.08 |
CALML4 |
calmodulin-like 4 |
69985 |
0.1 |
| chr3_167584284_167584435 | 0.08 |
SERPINI1 |
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
59333 |
0.15 |
| chr4_75550868_75551125 | 0.08 |
AC142293.3 |
|
36332 |
0.18 |
| chr6_116768830_116769011 | 0.08 |
ENSG00000265516 |
. |
10065 |
0.13 |
| chr8_125668530_125668681 | 0.08 |
MTSS1 |
metastasis suppressor 1 |
71252 |
0.1 |
| chr12_8671499_8671650 | 0.08 |
CLEC4D |
C-type lectin domain family 4, member D |
5438 |
0.16 |
| chr3_17062701_17062930 | 0.08 |
PLCL2 |
phospholipase C-like 2 |
10829 |
0.23 |
| chr13_100943012_100943163 | 0.08 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
2391 |
0.43 |
| chr10_6596246_6596398 | 0.08 |
PRKCQ |
protein kinase C, theta |
25879 |
0.26 |
| chr12_25213859_25214095 | 0.08 |
LRMP |
lymphoid-restricted membrane protein |
8303 |
0.21 |
| chr3_71890913_71891064 | 0.08 |
ENSG00000239250 |
. |
14660 |
0.23 |
| chr1_38317055_38317226 | 0.08 |
MTF1 |
metal-regulatory transcription factor 1 |
8152 |
0.1 |
| chr14_51294521_51294672 | 0.08 |
NIN |
ninein (GSK3B interacting protein) |
2601 |
0.2 |
| chr5_142702116_142702350 | 0.08 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
78184 |
0.11 |
| chr8_95023245_95023396 | 0.08 |
ENSG00000263855 |
. |
37238 |
0.18 |
| chr1_199059201_199059574 | 0.08 |
ENSG00000239006 |
. |
106925 |
0.08 |
| chr18_74236487_74236638 | 0.07 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
4050 |
0.22 |
| chr4_110621692_110621843 | 0.07 |
CASP6 |
caspase 6, apoptosis-related cysteine peptidase |
2238 |
0.28 |
| chr10_81951710_81952023 | 0.07 |
ANXA11 |
annexin A11 |
12381 |
0.2 |
| chr2_187362716_187362867 | 0.07 |
AC018867.1 |
Uncharacterized protein |
951 |
0.56 |
| chr15_58540473_58540733 | 0.07 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
30859 |
0.22 |
| chr21_34431974_34432125 | 0.07 |
OLIG1 |
oligodendrocyte transcription factor 1 |
10401 |
0.17 |
| chr2_158301630_158301829 | 0.07 |
CYTIP |
cytohesin 1 interacting protein |
1075 |
0.56 |
| chr11_5818563_5818714 | 0.07 |
OR52N1 |
olfactory receptor, family 52, subfamily N, member 1 |
8592 |
0.12 |
| chr3_183093543_183093694 | 0.07 |
MCF2L2 |
MCF.2 cell line derived transforming sequence-like 2 |
52147 |
0.1 |
| chr5_95161582_95161941 | 0.07 |
GLRX |
glutaredoxin (thioltransferase) |
3052 |
0.2 |
| chr2_24719419_24719570 | 0.07 |
NCOA1 |
nuclear receptor coactivator 1 |
4567 |
0.28 |
| chr1_118527197_118527348 | 0.07 |
SPAG17 |
sperm associated antigen 17 |
23421 |
0.21 |
| chr3_15258192_15258343 | 0.07 |
CAPN7 |
calpain 7 |
10517 |
0.16 |
| chr7_48293008_48293159 | 0.07 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
82026 |
0.11 |
| chr1_154687530_154687681 | 0.07 |
ADAR |
adenosine deaminase, RNA-specific |
87131 |
0.06 |
| chr3_172242674_172242825 | 0.07 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
1452 |
0.51 |
| chr11_7613293_7613444 | 0.07 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
5049 |
0.21 |
| chr12_59809371_59809522 | 0.07 |
ENSG00000251931 |
. |
35107 |
0.17 |
| chrX_100877158_100877309 | 0.07 |
ARMCX3 |
armadillo repeat containing, X-linked 3 |
554 |
0.58 |
| chr3_126178029_126178180 | 0.07 |
ZXDC |
ZXD family zinc finger C |
16604 |
0.16 |
| chr4_84161643_84161976 | 0.07 |
COQ2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
44109 |
0.16 |
| chr2_240876499_240876650 | 0.07 |
ENSG00000264279 |
. |
5937 |
0.25 |
| chrY_7156872_7157023 | 0.07 |
PRKY |
protein kinase, Y-linked, pseudogene |
14593 |
0.21 |
| chr14_68949048_68949199 | 0.07 |
RAD51B |
RAD51 paralog B |
70896 |
0.12 |
| chr15_76298943_76299094 | 0.07 |
NRG4 |
neuregulin 4 |
5400 |
0.23 |
| chrX_96479424_96479575 | 0.07 |
ENSG00000265260 |
. |
116546 |
0.07 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0051461 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0002691 | regulation of cellular extravasation(GO:0002691) positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.0 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.0 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0042723 | thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.0 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.0 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.0 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |