Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
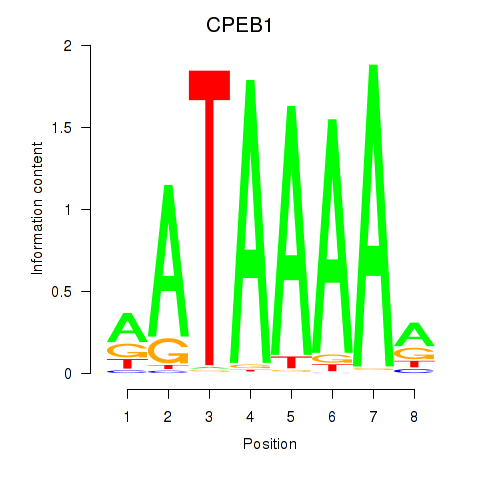
Results for CPEB1
Z-value: 1.30
Transcription factors associated with CPEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CPEB1
|
ENSG00000214575.5 | cytoplasmic polyadenylation element binding protein 1 |
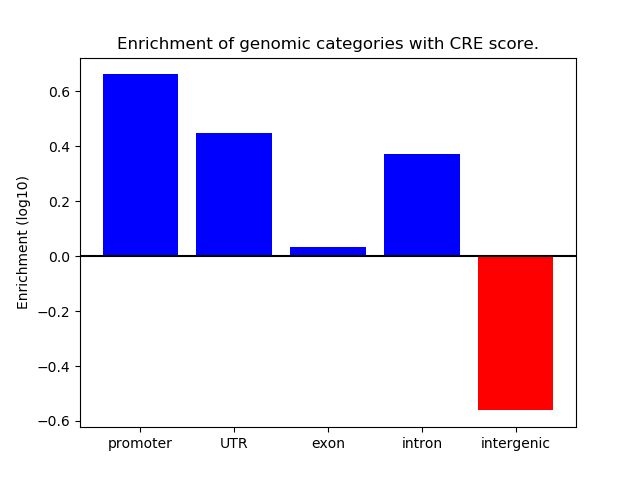
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_83315771_83316117 | CPEB1 | 70 | 0.898115 | 0.90 | 1.0e-03 | Click! |
| chr15_83239391_83239542 | CPEB1 | 1043 | 0.414979 | -0.61 | 8.0e-02 | Click! |
| chr15_83240171_83240322 | CPEB1 | 263 | 0.876035 | -0.53 | 1.4e-01 | Click! |
| chr15_83235277_83235428 | CPEB1 | 5157 | 0.136551 | -0.50 | 1.7e-01 | Click! |
| chr15_83241226_83241377 | CPEB1 | 473 | 0.732689 | -0.44 | 2.3e-01 | Click! |
Activity of the CPEB1 motif across conditions
Conditions sorted by the z-value of the CPEB1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
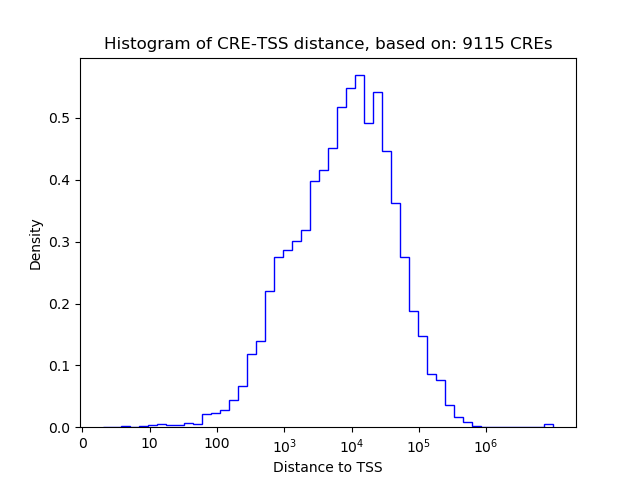
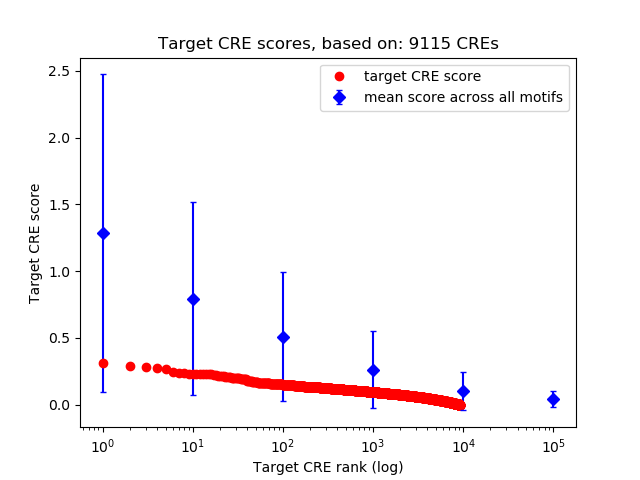
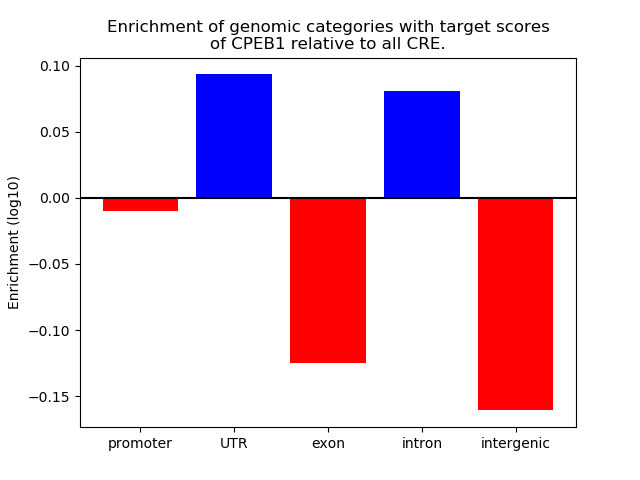
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_99721349_99721548 | 0.31 |
AL109767.1 |
|
7837 |
0.22 |
| chr17_37948646_37948906 | 0.29 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
14298 |
0.14 |
| chr17_47896409_47896901 | 0.28 |
RP11-304F15.3 |
|
26617 |
0.12 |
| chr10_22962552_22962703 | 0.28 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
40410 |
0.2 |
| chr10_43843752_43843981 | 0.27 |
ENSG00000221468 |
. |
6632 |
0.19 |
| chr2_143888983_143889411 | 0.25 |
ARHGAP15 |
Rho GTPase activating protein 15 |
2314 |
0.39 |
| chr20_56195788_56195967 | 0.24 |
ZBP1 |
Z-DNA binding protein 1 |
245 |
0.94 |
| chr8_2166897_2167149 | 0.24 |
MYOM2 |
myomesin 2 |
173839 |
0.03 |
| chr18_43808452_43808604 | 0.23 |
C18orf25 |
chromosome 18 open reading frame 25 |
54528 |
0.13 |
| chr9_115066997_115067151 | 0.23 |
PTBP3 |
polypyrimidine tract binding protein 3 |
28049 |
0.14 |
| chr17_33859475_33859650 | 0.23 |
SLFN12L |
schlafen family member 12-like |
5318 |
0.12 |
| chr14_92435696_92435980 | 0.23 |
FBLN5 |
fibulin 5 |
21507 |
0.18 |
| chr17_37934637_37934793 | 0.23 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
237 |
0.9 |
| chr3_114749732_114749883 | 0.23 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
40415 |
0.23 |
| chr3_18597997_18598194 | 0.23 |
ENSG00000228956 |
. |
29431 |
0.24 |
| chr3_197595834_197596031 | 0.23 |
LRCH3 |
leucine-rich repeats and calponin homology (CH) domain containing 3 |
2846 |
0.23 |
| chr10_32749220_32749452 | 0.22 |
CCDC7 |
coiled-coil domain containing 7 |
8787 |
0.22 |
| chr1_59144710_59144861 | 0.22 |
MYSM1 |
Myb-like, SWIRM and MPN domains 1 |
20979 |
0.2 |
| chrX_78517963_78518114 | 0.22 |
GPR174 |
G protein-coupled receptor 174 |
91569 |
0.09 |
| chr22_20919404_20919765 | 0.22 |
MED15 |
mediator complex subunit 15 |
14007 |
0.14 |
| chr9_95733021_95733387 | 0.21 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
3560 |
0.27 |
| chr14_35400951_35401102 | 0.21 |
ENSG00000199980 |
. |
1723 |
0.34 |
| chr11_108108099_108108300 | 0.21 |
ENSG00000206967 |
. |
7868 |
0.16 |
| chrX_53712364_53712515 | 0.21 |
HUWE1 |
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
1234 |
0.59 |
| chr3_151957728_151957952 | 0.21 |
MBNL1 |
muscleblind-like splicing regulator 1 |
27989 |
0.2 |
| chr1_193428730_193429035 | 0.21 |
ENSG00000252241 |
. |
272192 |
0.01 |
| chr22_29195216_29195505 | 0.20 |
XBP1 |
X-box binding protein 1 |
761 |
0.48 |
| chr6_89626872_89627052 | 0.20 |
RNGTT |
RNA guanylyltransferase and 5'-phosphatase |
46318 |
0.15 |
| chr17_62140117_62140268 | 0.20 |
ICAM2 |
intercellular adhesion molecule 2 |
42198 |
0.11 |
| chr14_99720519_99720759 | 0.20 |
AL109767.1 |
|
8646 |
0.22 |
| chr2_12849849_12850137 | 0.20 |
TRIB2 |
tribbles pseudokinase 2 |
7022 |
0.27 |
| chr2_238978929_238979080 | 0.20 |
SCLY |
selenocysteine lyase |
899 |
0.56 |
| chr11_60767130_60767281 | 0.20 |
CD6 |
CD6 molecule |
8801 |
0.13 |
| chr2_202059112_202059539 | 0.19 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
11407 |
0.15 |
| chr6_36077167_36077664 | 0.19 |
MAPK13 |
mitogen-activated protein kinase 13 |
20683 |
0.16 |
| chr11_5246485_5246763 | 0.19 |
ENSG00000221031 |
. |
755 |
0.38 |
| chr1_62188467_62188709 | 0.19 |
TM2D1 |
TM2 domain containing 1 |
2217 |
0.35 |
| chr5_178978355_178978506 | 0.19 |
RUFY1 |
RUN and FYVE domain containing 1 |
591 |
0.74 |
| chr16_9048805_9048966 | 0.19 |
USP7 |
ubiquitin specific peptidase 7 (herpes virus-associated) |
1352 |
0.47 |
| chr4_40189463_40189614 | 0.18 |
RHOH |
ras homolog family member H |
3135 |
0.27 |
| chr2_24272218_24272506 | 0.18 |
C2orf44 |
chromosome 2 open reading frame 44 |
83 |
0.67 |
| chr14_99691163_99691558 | 0.18 |
AL109767.1 |
|
37925 |
0.16 |
| chr2_106545238_106545692 | 0.18 |
AC009505.2 |
|
71832 |
0.11 |
| chr14_99726600_99726784 | 0.18 |
AL109767.1 |
|
2593 |
0.31 |
| chr2_177080_177288 | 0.17 |
AC079779.7 |
|
20385 |
0.21 |
| chr1_203259066_203259291 | 0.17 |
BTG2 |
BTG family, member 2 |
15486 |
0.16 |
| chr12_96755738_96755889 | 0.17 |
CDK17 |
cyclin-dependent kinase 17 |
26805 |
0.18 |
| chr17_8147401_8147552 | 0.17 |
PFAS |
phosphoribosylformylglycinamidine synthase |
3460 |
0.11 |
| chr1_101395582_101395733 | 0.17 |
SLC30A7 |
solute carrier family 30 (zinc transporter), member 7 |
31727 |
0.12 |
| chr12_66684119_66684270 | 0.17 |
ENSG00000222744 |
. |
2269 |
0.26 |
| chr4_143635559_143635710 | 0.17 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
131809 |
0.06 |
| chr14_61817217_61817623 | 0.17 |
PRKCH |
protein kinase C, eta |
2811 |
0.3 |
| chr14_21697616_21697899 | 0.17 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
1824 |
0.32 |
| chr17_46670095_46670275 | 0.17 |
HOXB-AS3 |
HOXB cluster antisense RNA 3 |
503 |
0.48 |
| chr12_42722388_42722539 | 0.16 |
PPHLN1 |
periphilin 1 |
2483 |
0.22 |
| chr18_60970002_60970404 | 0.16 |
RP11-28F1.2 |
|
11112 |
0.18 |
| chr3_60090466_60090753 | 0.16 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
133026 |
0.06 |
| chr8_66769940_66770103 | 0.16 |
PDE7A |
phosphodiesterase 7A |
15834 |
0.27 |
| chr7_8173692_8173968 | 0.16 |
AC006042.6 |
|
20175 |
0.19 |
| chr18_67626079_67626351 | 0.16 |
CD226 |
CD226 molecule |
1803 |
0.48 |
| chr20_57734207_57734542 | 0.16 |
ZNF831 |
zinc finger protein 831 |
31701 |
0.19 |
| chr3_133210635_133210930 | 0.16 |
BFSP2-AS1 |
BFSP2 antisense RNA 1 |
36162 |
0.16 |
| chr5_56031061_56031244 | 0.16 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
80249 |
0.09 |
| chr18_3259945_3260096 | 0.16 |
RP13-270P17.1 |
|
1800 |
0.26 |
| chr17_37925720_37925871 | 0.16 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
8683 |
0.13 |
| chr3_33843113_33843264 | 0.16 |
PDCD6IP |
programmed cell death 6 interacting protein |
3073 |
0.38 |
| chr7_12252889_12253205 | 0.16 |
TMEM106B |
transmembrane protein 106B |
2098 |
0.48 |
| chr15_26078408_26078559 | 0.16 |
ENSG00000199214 |
. |
15353 |
0.17 |
| chr17_48965132_48965283 | 0.16 |
TOB1 |
transducer of ERBB2, 1 |
19868 |
0.15 |
| chr14_99665484_99665650 | 0.16 |
AL162151.4 |
|
40814 |
0.17 |
| chr2_204596528_204596679 | 0.16 |
CD28 |
CD28 molecule |
25187 |
0.19 |
| chr7_135347931_135348131 | 0.16 |
C7orf73 |
chromosome 7 open reading frame 73 |
762 |
0.59 |
| chr16_67097233_67097384 | 0.16 |
CBFB |
core-binding factor, beta subunit |
33453 |
0.09 |
| chr1_101396601_101396752 | 0.16 |
SLC30A7 |
solute carrier family 30 (zinc transporter), member 7 |
30708 |
0.13 |
| chr2_38056365_38056516 | 0.16 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
90829 |
0.08 |
| chr12_25536580_25536731 | 0.16 |
ENSG00000201439 |
. |
20707 |
0.24 |
| chr17_26206098_26206388 | 0.16 |
LYRM9 |
LYR motif containing 9 |
4080 |
0.19 |
| chr5_54391771_54392054 | 0.16 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
6564 |
0.16 |
| chr3_33038134_33038326 | 0.16 |
CCR4 |
chemokine (C-C motif) receptor 4 |
45164 |
0.15 |
| chr16_12006362_12006679 | 0.16 |
GSPT1 |
G1 to S phase transition 1 |
2540 |
0.15 |
| chr14_98637494_98637668 | 0.16 |
ENSG00000222066 |
. |
160506 |
0.04 |
| chr6_112006099_112006376 | 0.16 |
FYN |
FYN oncogene related to SRC, FGR, YES |
35028 |
0.16 |
| chr5_55391166_55391317 | 0.16 |
ANKRD55 |
ankyrin repeat domain 55 |
21537 |
0.16 |
| chr5_148733548_148733699 | 0.16 |
GRPEL2-AS1 |
GRPEL2 atnisense RNA 1 |
3582 |
0.14 |
| chr3_46411260_46411477 | 0.16 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
265 |
0.9 |
| chr1_8212069_8212240 | 0.15 |
ENSG00000200975 |
. |
54503 |
0.13 |
| chr13_40763533_40763684 | 0.15 |
ENSG00000207458 |
. |
37356 |
0.23 |
| chr17_71208357_71208949 | 0.15 |
COG1 |
component of oligomeric golgi complex 1 |
6355 |
0.15 |
| chr2_204812325_204812548 | 0.15 |
ICOS |
inducible T-cell co-stimulator |
10933 |
0.28 |
| chr6_111495332_111495527 | 0.15 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
1531 |
0.44 |
| chr12_9807704_9808028 | 0.15 |
RP11-705C15.2 |
|
1587 |
0.25 |
| chr10_120458675_120458868 | 0.15 |
CACUL1 |
CDK2-associated, cullin domain 1 |
55987 |
0.12 |
| chr4_143621409_143621560 | 0.15 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
139662 |
0.05 |
| chr13_100194616_100194885 | 0.15 |
ENSG00000212197 |
. |
6100 |
0.2 |
| chr2_152144415_152144634 | 0.15 |
NMI |
N-myc (and STAT) interactor |
1884 |
0.36 |
| chr6_151735099_151735250 | 0.15 |
ZBTB2 |
zinc finger and BTB domain containing 2 |
22491 |
0.13 |
| chr22_22117730_22117960 | 0.15 |
YPEL1 |
yippee-like 1 (Drosophila) |
27722 |
0.11 |
| chr2_44796225_44796376 | 0.15 |
CAMKMT |
calmodulin-lysine N-methyltransferase |
196419 |
0.03 |
| chr12_22484745_22484973 | 0.15 |
ST8SIA1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
2238 |
0.44 |
| chr14_38546372_38546618 | 0.15 |
CTD-2058B24.2 |
|
13868 |
0.25 |
| chr7_130596773_130597211 | 0.15 |
ENSG00000226380 |
. |
34694 |
0.19 |
| chr6_139485972_139486167 | 0.15 |
HECA |
headcase homolog (Drosophila) |
29820 |
0.2 |
| chr2_10815541_10815692 | 0.15 |
NOL10 |
nucleolar protein 10 |
3800 |
0.22 |
| chr6_502144_502346 | 0.15 |
RP1-20B11.2 |
|
21926 |
0.26 |
| chr2_158746528_158746679 | 0.15 |
UPP2 |
uridine phosphorylase 2 |
13389 |
0.16 |
| chr2_95741879_95742163 | 0.15 |
AC103563.9 |
|
23100 |
0.15 |
| chr1_167458272_167458474 | 0.15 |
CD247 |
CD247 molecule |
29402 |
0.16 |
| chr22_40296066_40296492 | 0.15 |
GRAP2 |
GRB2-related adaptor protein 2 |
807 |
0.6 |
| chrX_12972541_12972692 | 0.15 |
TMSB4X |
thymosin beta 4, X-linked |
20611 |
0.19 |
| chr12_45269447_45269715 | 0.15 |
NELL2 |
NEL-like 2 (chicken) |
14 |
0.98 |
| chr4_82414160_82414311 | 0.15 |
RASGEF1B |
RasGEF domain family, member 1B |
21166 |
0.28 |
| chr2_102186414_102186691 | 0.15 |
RFX8 |
RFX family member 8, lacking RFX DNA binding domain |
95387 |
0.08 |
| chr14_98665522_98665718 | 0.15 |
ENSG00000222066 |
. |
132467 |
0.05 |
| chr8_126940986_126941137 | 0.15 |
ENSG00000206695 |
. |
27866 |
0.26 |
| chrX_70763186_70763395 | 0.15 |
OGT |
O-linked N-acetylglucosamine (GlcNAc) transferase |
6426 |
0.2 |
| chr3_56912022_56912345 | 0.15 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
38316 |
0.18 |
| chr11_133798697_133798899 | 0.15 |
IGSF9B |
immunoglobulin superfamily, member 9B |
17389 |
0.18 |
| chr10_11252064_11252436 | 0.15 |
RP3-323N1.2 |
|
38911 |
0.17 |
| chr6_143862532_143862683 | 0.15 |
PHACTR2 |
phosphatase and actin regulator 2 |
4625 |
0.18 |
| chr17_33171161_33171312 | 0.15 |
CCT6B |
chaperonin containing TCP1, subunit 6B (zeta 2) |
117185 |
0.05 |
| chr5_172425652_172425803 | 0.15 |
ATP6V0E1 |
ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 |
14893 |
0.14 |
| chr2_7052046_7052272 | 0.14 |
RNF144A |
ring finger protein 144A |
5364 |
0.19 |
| chr1_77774898_77775049 | 0.14 |
AK5 |
adenylate kinase 5 |
26209 |
0.18 |
| chr1_111326257_111326605 | 0.14 |
ENSG00000199710 |
. |
19303 |
0.2 |
| chr11_117916276_117916450 | 0.14 |
ENSG00000272075 |
. |
21589 |
0.14 |
| chr1_184813678_184813906 | 0.14 |
ENSG00000252222 |
. |
23169 |
0.2 |
| chr11_35351604_35352020 | 0.14 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
28737 |
0.17 |
| chr5_156953479_156953635 | 0.14 |
ADAM19 |
ADAM metallopeptidase domain 19 |
23727 |
0.15 |
| chr5_39190224_39190381 | 0.14 |
FYB |
FYN binding protein |
12827 |
0.27 |
| chr11_108084515_108084666 | 0.14 |
ATM |
ataxia telangiectasia mutated |
8621 |
0.16 |
| chr3_151929579_151929730 | 0.14 |
MBNL1 |
muscleblind-like splicing regulator 1 |
56175 |
0.14 |
| chr21_16893074_16893225 | 0.14 |
ENSG00000212564 |
. |
93453 |
0.09 |
| chr14_22621058_22621209 | 0.14 |
ENSG00000238634 |
. |
10246 |
0.28 |
| chr3_101832900_101833053 | 0.14 |
ZPLD1 |
zona pellucida-like domain containing 1 |
14888 |
0.29 |
| chr13_42041149_42041395 | 0.14 |
RGCC |
regulator of cell cycle |
9577 |
0.16 |
| chr1_121311422_121311588 | 0.14 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
375568 |
0.01 |
| chr6_110882269_110882528 | 0.14 |
CDK19 |
cyclin-dependent kinase 19 |
82123 |
0.08 |
| chr14_64329252_64329512 | 0.14 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
9650 |
0.22 |
| chr3_195250896_195251062 | 0.14 |
ENSG00000252620 |
. |
960 |
0.52 |
| chr4_40236655_40236806 | 0.14 |
RHOH |
ras homolog family member H |
34766 |
0.17 |
| chr1_93420237_93420388 | 0.14 |
FAM69A |
family with sequence similarity 69, member A |
6745 |
0.13 |
| chr16_53530038_53530189 | 0.14 |
AKTIP |
AKT interacting protein |
4128 |
0.23 |
| chr1_43500743_43500945 | 0.14 |
ENSG00000252803 |
. |
11536 |
0.19 |
| chr4_148972762_148973133 | 0.14 |
RP11-76G10.1 |
|
94675 |
0.09 |
| chr5_14704726_14704885 | 0.14 |
FAM105B |
family with sequence similarity 105, member B |
23146 |
0.19 |
| chr6_35649971_35650157 | 0.14 |
FKBP5 |
FK506 binding protein 5 |
6628 |
0.14 |
| chr3_16356399_16356550 | 0.14 |
RP11-415F23.2 |
|
528 |
0.76 |
| chr2_205836626_205836826 | 0.14 |
PARD3B |
par-3 family cell polarity regulator beta |
426003 |
0.01 |
| chr17_37946299_37946450 | 0.14 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
11896 |
0.14 |
| chr14_21136496_21136717 | 0.14 |
ENSG00000199461 |
. |
10964 |
0.09 |
| chr22_40730411_40730669 | 0.14 |
ADSL |
adenylosuccinate lyase |
11967 |
0.19 |
| chr10_124175104_124175255 | 0.14 |
ENSG00000265442 |
. |
1302 |
0.43 |
| chr6_130505643_130505870 | 0.14 |
SAMD3 |
sterile alpha motif domain containing 3 |
29758 |
0.2 |
| chr15_57857934_57858132 | 0.14 |
GCOM1 |
GRINL1A complex locus 1 |
26073 |
0.17 |
| chr19_1057848_1057999 | 0.14 |
ABCA7 |
ATP-binding cassette, sub-family A (ABC1), member 7 |
990 |
0.32 |
| chr14_102294369_102294541 | 0.14 |
CTD-2017C7.1 |
|
11413 |
0.15 |
| chr3_15578936_15579113 | 0.14 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
15766 |
0.14 |
| chr7_129545877_129546028 | 0.14 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
45215 |
0.1 |
| chr13_27296341_27296492 | 0.14 |
GPR12 |
G protein-coupled receptor 12 |
38011 |
0.2 |
| chr7_142504543_142504845 | 0.14 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
23563 |
0.15 |
| chr18_13297039_13297305 | 0.14 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
19070 |
0.18 |
| chr1_153234965_153235116 | 0.14 |
LOR |
loricrin |
2864 |
0.19 |
| chr6_110515654_110515805 | 0.14 |
CDC40 |
cell division cycle 40 |
14105 |
0.22 |
| chr6_36932436_36932655 | 0.14 |
PI16 |
peptidase inhibitor 16 |
10336 |
0.17 |
| chr4_143621654_143621881 | 0.14 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
139945 |
0.05 |
| chr14_90105259_90105410 | 0.14 |
RP11-944C7.1 |
Protein LOC100506792 |
7424 |
0.15 |
| chr12_45608916_45609133 | 0.14 |
ANO6 |
anoctamin 6 |
779 |
0.77 |
| chr6_26358903_26359054 | 0.14 |
ENSG00000252399 |
. |
5714 |
0.1 |
| chr6_139492633_139492784 | 0.14 |
HECA |
headcase homolog (Drosophila) |
36459 |
0.17 |
| chr14_106630342_106630519 | 0.14 |
IGHV1-17 |
immunoglobulin heavy variable 1-17 (pseudogene) |
1019 |
0.19 |
| chr17_46508666_46508817 | 0.14 |
SKAP1 |
src kinase associated phosphoprotein 1 |
1160 |
0.41 |
| chr16_28998091_28998242 | 0.14 |
LAT |
linker for activation of T cells |
633 |
0.49 |
| chr1_24839011_24839397 | 0.14 |
RCAN3 |
RCAN family member 3 |
1606 |
0.35 |
| chr6_108109631_108109782 | 0.14 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
16251 |
0.26 |
| chr16_67538677_67538891 | 0.14 |
FAM65A |
family with sequence similarity 65, member A |
13537 |
0.09 |
| chr10_63809503_63809654 | 0.14 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
608 |
0.82 |
| chr10_90590706_90591168 | 0.14 |
ANKRD22 |
ankyrin repeat domain 22 |
20638 |
0.15 |
| chr15_40743963_40744180 | 0.14 |
RP11-64K12.9 |
|
2767 |
0.14 |
| chr10_14605059_14605230 | 0.14 |
FAM107B |
family with sequence similarity 107, member B |
6965 |
0.26 |
| chr1_90077786_90078098 | 0.14 |
RP11-413E1.4 |
|
17342 |
0.16 |
| chr5_80849712_80849863 | 0.14 |
SSBP2 |
single-stranded DNA binding protein 2 |
40251 |
0.21 |
| chr4_110496140_110496528 | 0.13 |
CCDC109B |
coiled-coil domain containing 109B |
14188 |
0.22 |
| chr10_6635069_6635386 | 0.13 |
PRKCQ |
protein kinase C, theta |
12964 |
0.3 |
| chr2_95690474_95690625 | 0.13 |
MAL |
mal, T-cell differentiation protein |
880 |
0.49 |
| chr15_75725504_75725655 | 0.13 |
SIN3A |
SIN3 transcription regulator family member A |
18347 |
0.15 |
| chr8_101728028_101728286 | 0.13 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
1961 |
0.25 |
| chr8_29389067_29389218 | 0.13 |
RP4-676L2.1 |
|
178455 |
0.03 |
| chr17_76710387_76710538 | 0.13 |
CYTH1 |
cytohesin 1 |
2645 |
0.28 |
| chr10_52380207_52380358 | 0.13 |
SGMS1 |
sphingomyelin synthase 1 |
3461 |
0.21 |
| chr19_11085110_11085263 | 0.13 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
9642 |
0.13 |
| chr1_7811961_7812165 | 0.13 |
CAMTA1 |
calmodulin binding transcription activator 1 |
7042 |
0.17 |
| chr21_43827327_43827478 | 0.13 |
UBASH3A |
ubiquitin associated and SH3 domain containing A |
3376 |
0.17 |
| chr20_52216616_52216767 | 0.13 |
ZNF217 |
zinc finger protein 217 |
6313 |
0.21 |
| chr2_12641093_12641393 | 0.13 |
ENSG00000207183 |
. |
89616 |
0.1 |
| chr14_98606379_98606534 | 0.13 |
C14orf64 |
chromosome 14 open reading frame 64 |
161995 |
0.04 |
| chr9_74375544_74375695 | 0.13 |
TMEM2 |
transmembrane protein 2 |
7683 |
0.3 |
| chr5_54399188_54399417 | 0.13 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
826 |
0.55 |
| chr5_118574777_118574928 | 0.13 |
ENSG00000266587 |
. |
18280 |
0.18 |
| chr5_49931517_49931668 | 0.13 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
30141 |
0.26 |
| chr1_100856626_100856777 | 0.13 |
ENSG00000216067 |
. |
12370 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.3 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.2 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.1 | 0.2 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.1 | GO:1903312 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.1 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.2 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.2 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0002363 | alpha-beta T cell lineage commitment(GO:0002363) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.2 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 1.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0002837 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0033083 | regulation of immature T cell proliferation(GO:0033083) regulation of immature T cell proliferation in thymus(GO:0033084) |
| 0.0 | 0.0 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.1 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.0 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.0 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.0 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0022009 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.0 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0002707 | negative regulation of lymphocyte mediated immunity(GO:0002707) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.0 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.0 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:0002704 | negative regulation of leukocyte mediated immunity(GO:0002704) |
| 0.0 | 0.0 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.0 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.0 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.0 | 0.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.0 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.0 | 0.0 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.0 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.2 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.0 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.6 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.0 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |