Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
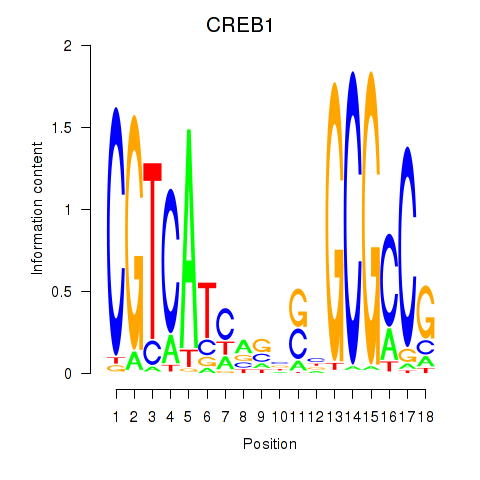
Results for CREB1
Z-value: 0.64
Transcription factors associated with CREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB1
|
ENSG00000118260.10 | cAMP responsive element binding protein 1 |
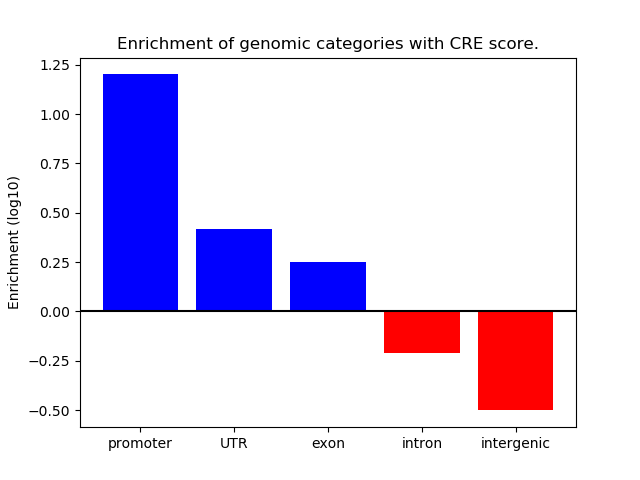
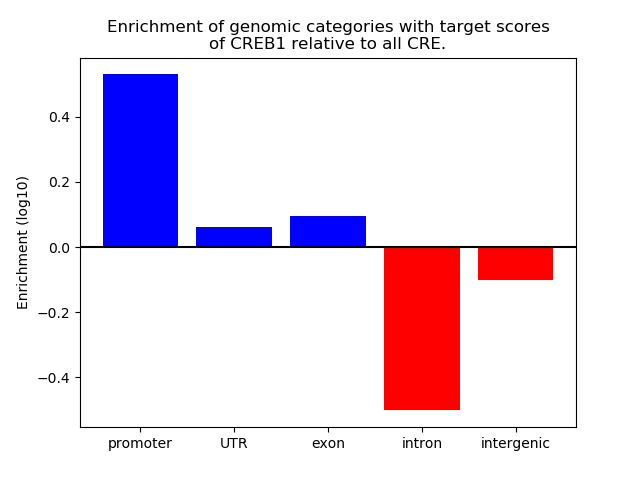
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_208394003_208394154 | CREB1 | 383 | 0.525119 | 0.75 | 1.9e-02 | Click! |
| chr2_208396639_208396800 | CREB1 | 1891 | 0.318398 | 0.73 | 2.6e-02 | Click! |
| chr2_208413656_208413807 | CREB1 | 1254 | 0.492185 | -0.69 | 3.8e-02 | Click! |
| chr2_208421027_208421178 | CREB1 | 2744 | 0.288571 | -0.67 | 5.1e-02 | Click! |
| chr2_208411969_208412295 | CREB1 | 2853 | 0.282219 | -0.65 | 5.6e-02 | Click! |
Activity of the CREB1 motif across conditions
Conditions sorted by the z-value of the CREB1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
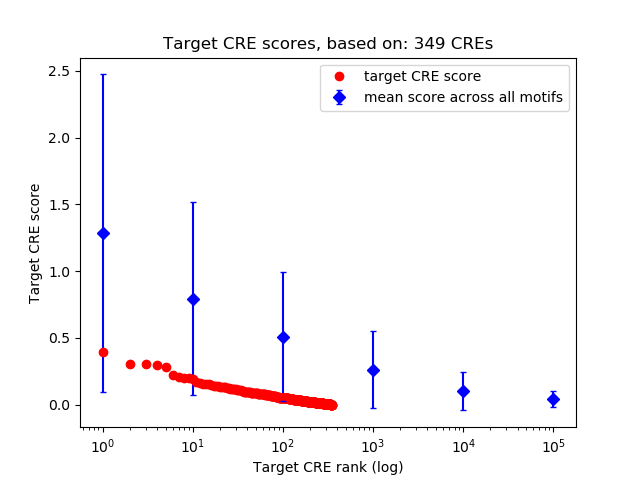
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_40014894_40015175 | 0.40 |
BCOR |
BCL6 corepressor |
9152 |
0.31 |
| chr18_77138968_77139119 | 0.31 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
16813 |
0.2 |
| chr22_41681891_41682042 | 0.30 |
RANGAP1 |
Ran GTPase activating protein 1 |
3 |
0.96 |
| chr1_27191187_27191395 | 0.30 |
SFN |
stratifin |
1658 |
0.22 |
| chr12_54145261_54145434 | 0.29 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
23818 |
0.14 |
| chr11_133907189_133907404 | 0.22 |
JAM3 |
junctional adhesion molecule 3 |
31524 |
0.18 |
| chr2_216877823_216877974 | 0.21 |
MREG |
melanoregulin |
448 |
0.87 |
| chr13_42615279_42615552 | 0.20 |
DGKH |
diacylglycerol kinase, eta |
1239 |
0.48 |
| chr3_32022334_32022485 | 0.20 |
OSBPL10 |
oxysterol binding protein-like 10 |
383 |
0.73 |
| chr1_64059758_64059909 | 0.19 |
PGM1 |
phosphoglucomutase 1 |
353 |
0.87 |
| chr7_149411412_149411638 | 0.17 |
KRBA1 |
KRAB-A domain containing 1 |
347 |
0.9 |
| chr6_52463129_52463280 | 0.16 |
TRAM2 |
translocation associated membrane protein 2 |
21491 |
0.21 |
| chr9_96793340_96793664 | 0.16 |
PTPDC1 |
protein tyrosine phosphatase domain containing 1 |
426 |
0.88 |
| chr7_12443468_12443619 | 0.15 |
VWDE |
von Willebrand factor D and EGF domains |
12 |
0.99 |
| chr20_3088467_3088718 | 0.15 |
UBOX5-AS1 |
UBOX5 antisense RNA 1 |
1033 |
0.39 |
| chr4_8430147_8430298 | 0.14 |
ACOX3 |
acyl-CoA oxidase 3, pristanoyl |
9 |
0.98 |
| chr12_54350090_54350429 | 0.14 |
HOXC12 |
homeobox C12 |
1641 |
0.16 |
| chr19_8008904_8009202 | 0.14 |
TIMM44 |
translocase of inner mitochondrial membrane 44 homolog (yeast) |
248 |
0.81 |
| chr12_133022629_133022780 | 0.14 |
MUC8 |
mucin 8 |
28022 |
0.17 |
| chr2_240386919_240387070 | 0.14 |
AC062017.1 |
Uncharacterized protein |
63495 |
0.11 |
| chr9_99800693_99800974 | 0.13 |
CTSV |
cathepsin V |
759 |
0.71 |
| chr18_74773774_74774062 | 0.13 |
MBP |
myelin basic protein |
43299 |
0.18 |
| chr1_109643083_109643234 | 0.13 |
ENSG00000270066 |
. |
343 |
0.8 |
| chr6_109775900_109776051 | 0.13 |
MICAL1 |
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
950 |
0.41 |
| chr12_52569726_52569877 | 0.12 |
KRT80 |
keratin 80 |
15983 |
0.12 |
| chr15_45353267_45353418 | 0.12 |
RP11-109D20.1 |
|
2437 |
0.16 |
| chr10_103577270_103577421 | 0.12 |
MGEA5 |
meningioma expressed antigen 5 (hyaluronidase) |
278 |
0.85 |
| chr16_30007199_30007350 | 0.12 |
INO80E |
INO80 complex subunit E |
238 |
0.53 |
| chr5_70751007_70751459 | 0.12 |
BDP1 |
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
209 |
0.96 |
| chr1_19970427_19970607 | 0.12 |
NBL1 |
neuroblastoma 1, DAN family BMP antagonist |
140 |
0.95 |
| chr17_78796891_78797091 | 0.11 |
RP11-28G8.1 |
|
17559 |
0.21 |
| chr10_121411607_121411991 | 0.11 |
BAG3 |
BCL2-associated athanogene 3 |
917 |
0.64 |
| chr4_6326348_6326522 | 0.11 |
WFS1 |
Wolfram syndrome 1 (wolframin) |
35633 |
0.19 |
| chr11_9595375_9595526 | 0.11 |
WEE1 |
WEE1 G2 checkpoint kinase |
222 |
0.91 |
| chr10_130008845_130008996 | 0.10 |
MKI67 |
marker of proliferation Ki-67 |
84271 |
0.11 |
| chr19_8454744_8454908 | 0.10 |
RAB11B |
RAB11B, member RAS oncogene family |
39 |
0.64 |
| chr22_24321820_24321971 | 0.10 |
DDT |
D-dopachrome tautomerase |
124 |
0.8 |
| chr4_8862024_8862175 | 0.10 |
RP13-582L3.4 |
|
454 |
0.85 |
| chr1_3075630_3075886 | 0.10 |
RP1-163G9.2 |
|
25485 |
0.21 |
| chr1_205426191_205426365 | 0.10 |
LEMD1 |
LEM domain containing 1 |
7219 |
0.17 |
| chr9_34589972_34590123 | 0.10 |
CNTFR |
ciliary neurotrophic factor receptor |
74 |
0.94 |
| chr2_121199656_121199846 | 0.09 |
LINC01101 |
long intergenic non-protein coding RNA 1101 |
23946 |
0.24 |
| chr5_6633870_6634021 | 0.09 |
SRD5A1 |
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
364 |
0.63 |
| chr3_156544046_156544197 | 0.09 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
18 |
0.99 |
| chr3_15160267_15160418 | 0.09 |
ENSG00000221424 |
. |
15695 |
0.16 |
| chr11_63767165_63767355 | 0.09 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
12946 |
0.11 |
| chr5_176734587_176734738 | 0.09 |
PRELID1 |
PRELI domain containing 1 |
2973 |
0.14 |
| chr1_246230838_246231074 | 0.09 |
RP11-83A16.1 |
|
34103 |
0.18 |
| chr2_45397824_45397975 | 0.09 |
SIX2 |
SIX homeobox 2 |
161330 |
0.04 |
| chr13_113764056_113764207 | 0.09 |
F7 |
coagulation factor VII (serum prothrombin conversion accelerator) |
4010 |
0.14 |
| chr3_66023505_66023773 | 0.09 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
344 |
0.9 |
| chr10_52382609_52382760 | 0.09 |
SGMS1 |
sphingomyelin synthase 1 |
1059 |
0.45 |
| chr16_3207501_3207863 | 0.08 |
CASP16 |
caspase 16, apoptosis-related cysteine peptidase (putative) |
13438 |
0.08 |
| chr12_104682747_104683040 | 0.08 |
TXNRD1 |
thioredoxin reductase 1 |
220 |
0.93 |
| chr1_1135456_1135661 | 0.08 |
TNFRSF18 |
tumor necrosis factor receptor superfamily, member 18 |
5502 |
0.08 |
| chr16_75551132_75551283 | 0.08 |
RP11-77K12.5 |
|
12026 |
0.1 |
| chr9_139426377_139426528 | 0.08 |
RP11-413M3.4 |
|
10881 |
0.09 |
| chr1_6661858_6662009 | 0.08 |
KLHL21 |
kelch-like family member 21 |
996 |
0.41 |
| chr13_103451004_103451240 | 0.08 |
KDELC1 |
KDEL (Lys-Asp-Glu-Leu) containing 1 |
235 |
0.52 |
| chr20_58004406_58004557 | 0.08 |
ENSG00000263903 |
. |
82946 |
0.1 |
| chr1_198127223_198127697 | 0.08 |
NEK7 |
NIMA-related kinase 7 |
1209 |
0.65 |
| chr16_2255933_2256271 | 0.08 |
MLST8 |
MTOR associated protein, LST8 homolog (S. cerevisiae) |
257 |
0.7 |
| chr22_50765742_50765893 | 0.08 |
DENND6B |
DENN/MADD domain containing 6B |
328 |
0.78 |
| chr4_8859377_8859610 | 0.08 |
RP13-582L3.4 |
|
3060 |
0.29 |
| chr11_112832512_112832665 | 0.08 |
NCAM1 |
neural cell adhesion molecule 1 |
386 |
0.61 |
| chr2_91997601_91997765 | 0.07 |
IGKV1OR-1 |
immunoglobulin kappa variable 1/OR-1 (pseudogene) |
8114 |
0.25 |
| chr1_27895325_27895557 | 0.07 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
34661 |
0.12 |
| chr3_69591941_69592092 | 0.07 |
FRMD4B |
FERM domain containing 4B |
282 |
0.95 |
| chr3_67704618_67704769 | 0.07 |
SUCLG2 |
succinate-CoA ligase, GDP-forming, beta subunit |
338 |
0.94 |
| chr21_32930330_32930496 | 0.07 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
877 |
0.48 |
| chr19_56592075_56592344 | 0.07 |
ZNF787 |
zinc finger protein 787 |
22644 |
0.12 |
| chr2_60962980_60963131 | 0.07 |
ENSG00000252718 |
. |
16280 |
0.19 |
| chr3_183735605_183735756 | 0.07 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
3 |
0.96 |
| chr17_17626155_17626306 | 0.07 |
RAI1 |
retinoic acid induced 1 |
40421 |
0.11 |
| chr6_97285605_97285766 | 0.07 |
GPR63 |
G protein-coupled receptor 63 |
332 |
0.91 |
| chr2_152494016_152494167 | 0.07 |
NEB |
nebulin |
53958 |
0.17 |
| chr1_46954889_46955040 | 0.07 |
DMBX1 |
diencephalon/mesencephalon homeobox 1 |
17704 |
0.16 |
| chr12_26111020_26111411 | 0.07 |
RASSF8-AS1 |
RASSF8 atnisense RNA 1 |
236 |
0.84 |
| chr5_134375155_134375381 | 0.06 |
PITX1 |
paired-like homeodomain 1 |
4765 |
0.22 |
| chr17_7464631_7464910 | 0.06 |
SENP3 |
SUMO1/sentrin/SMT3 specific peptidase 3 |
502 |
0.46 |
| chr17_80709161_80709321 | 0.06 |
TBCD |
tubulin folding cofactor D |
699 |
0.52 |
| chr13_110437392_110437543 | 0.06 |
IRS2 |
insulin receptor substrate 2 |
1448 |
0.54 |
| chr16_11295567_11296301 | 0.06 |
RMI2 |
RecQ mediated genome instability 2 |
47572 |
0.08 |
| chr10_50604077_50604326 | 0.06 |
DRGX |
dorsal root ganglia homeobox |
704 |
0.7 |
| chr17_5000284_5000552 | 0.06 |
ENSG00000234327 |
. |
14345 |
0.09 |
| chr7_117854342_117854690 | 0.06 |
ANKRD7 |
ankyrin repeat domain 7 |
10196 |
0.26 |
| chr17_46675290_46675535 | 0.06 |
HOXB-AS3 |
HOXB cluster antisense RNA 3 |
1611 |
0.15 |
| chr14_67708458_67708609 | 0.06 |
MPP5 |
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
189 |
0.95 |
| chr14_100906069_100906661 | 0.05 |
RP11-362L22.1 |
|
33123 |
0.1 |
| chr17_43096938_43097182 | 0.05 |
DCAKD |
dephospho-CoA kinase domain containing |
15449 |
0.12 |
| chr4_76861765_76862107 | 0.05 |
NAAA |
N-acylethanolamine acid amidase |
190 |
0.92 |
| chr6_43394965_43395176 | 0.05 |
ABCC10 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
34 |
0.96 |
| chr21_28339316_28339602 | 0.05 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
627 |
0.77 |
| chr22_20790969_20791236 | 0.05 |
SCARF2 |
scavenger receptor class F, member 2 |
1010 |
0.39 |
| chr21_39755799_39755950 | 0.05 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
87026 |
0.09 |
| chr17_79374508_79374888 | 0.05 |
ENSG00000266392 |
. |
120 |
0.91 |
| chr17_40215803_40215954 | 0.05 |
ZNF385C |
zinc finger protein 385C |
803 |
0.47 |
| chr5_176918444_176918625 | 0.05 |
RP11-1334A24.6 |
|
3462 |
0.12 |
| chr10_112404047_112404198 | 0.05 |
RBM20 |
RNA binding motif protein 20 |
33 |
0.96 |
| chr10_94449168_94449319 | 0.05 |
HHEX |
hematopoietically expressed homeobox |
1298 |
0.46 |
| chr1_233463868_233464019 | 0.05 |
MLK4 |
Mitogen-activated protein kinase kinase kinase MLK4 |
429 |
0.88 |
| chr15_44093066_44093217 | 0.05 |
HYPK |
huntingtin interacting protein K |
344 |
0.48 |
| chr4_140477398_140477823 | 0.05 |
SETD7 |
SET domain containing (lysine methyltransferase) 7 |
218 |
0.64 |
| chr1_166890270_166890421 | 0.05 |
TADA1 |
transcriptional adaptor 1 |
44781 |
0.12 |
| chr20_388183_388334 | 0.05 |
RBCK1 |
RanBP-type and C3HC4-type zinc finger containing 1 |
116 |
0.95 |
| chr9_136024815_136024979 | 0.05 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
176 |
0.93 |
| chr2_61696354_61697036 | 0.05 |
USP34 |
ubiquitin specific peptidase 34 |
1209 |
0.39 |
| chr1_230561180_230561454 | 0.05 |
PGBD5 |
piggyBac transposable element derived 5 |
158 |
0.97 |
| chr1_11750833_11751136 | 0.05 |
MAD2L2 |
MAD2 mitotic arrest deficient-like 2 (yeast) |
550 |
0.51 |
| chr13_26625030_26625181 | 0.05 |
SHISA2 |
shisa family member 2 |
64 |
0.98 |
| chr11_85565948_85566099 | 0.05 |
AP000974.1 |
CDNA FLJ26432 fis, clone KDN01418; Uncharacterized protein |
37 |
0.6 |
| chr19_49925689_49925950 | 0.05 |
PTH2 |
parathyroid hormone 2 |
879 |
0.27 |
| chr11_63766456_63766677 | 0.05 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
12252 |
0.11 |
| chr14_21100640_21100876 | 0.05 |
OR6S1 |
olfactory receptor, family 6, subfamily S, member 1 |
9092 |
0.1 |
| chr11_61355500_61355795 | 0.05 |
SYT7 |
synaptotagmin VII |
7027 |
0.19 |
| chr21_36261605_36261866 | 0.05 |
RUNX1 |
runt-related transcription factor 1 |
299 |
0.95 |
| chr1_47973634_47973787 | 0.05 |
ENSG00000221346 |
. |
5534 |
0.24 |
| chr20_7999916_8000067 | 0.04 |
RP5-971N18.3 |
|
121 |
0.84 |
| chr10_35931971_35932320 | 0.04 |
FZD8 |
frizzled family receptor 8 |
1783 |
0.3 |
| chr9_140008930_140009195 | 0.04 |
DPP7 |
dipeptidyl-peptidase 7 |
94 |
0.9 |
| chr20_24961168_24961319 | 0.04 |
ENSG00000207355 |
. |
6436 |
0.19 |
| chr15_81426559_81426733 | 0.04 |
C15orf26 |
chromosome 15 open reading frame 26 |
58 |
0.98 |
| chr2_28615239_28615407 | 0.04 |
FOSL2 |
FOS-like antigen 2 |
346 |
0.84 |
| chr18_53447366_53447551 | 0.04 |
TCF4 |
transcription factor 4 |
115440 |
0.07 |
| chr12_123716874_123717156 | 0.04 |
C12orf65 |
chromosome 12 open reading frame 65 |
448 |
0.55 |
| chr5_76506800_76506951 | 0.04 |
PDE8B |
phosphodiesterase 8B |
124 |
0.97 |
| chr7_151441431_151441582 | 0.04 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
8106 |
0.24 |
| chr9_79637084_79637292 | 0.04 |
FOXB2 |
forkhead box B2 |
2617 |
0.4 |
| chr6_116792861_116793073 | 0.04 |
RP1-93H18.6 |
|
8112 |
0.12 |
| chr6_31632946_31633178 | 0.04 |
CSNK2B |
casein kinase 2, beta polypeptide |
49 |
0.51 |
| chr1_205257351_205257502 | 0.04 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
32097 |
0.12 |
| chr3_123167522_123167831 | 0.04 |
ADCY5 |
adenylate cyclase 5 |
929 |
0.67 |
| chr5_43042322_43042506 | 0.04 |
AC025171.1 |
|
555 |
0.55 |
| chr10_12392184_12392335 | 0.04 |
CAMK1D |
calcium/calmodulin-dependent protein kinase ID |
527 |
0.78 |
| chr2_106350382_106350589 | 0.04 |
NCK2 |
NCK adaptor protein 2 |
10869 |
0.3 |
| chr17_66130072_66130259 | 0.04 |
LRRC37A16P |
leucine rich repeat containing 37, member A16, pseudogene |
18444 |
0.18 |
| chr2_231692888_231693039 | 0.04 |
ITM2C |
integral membrane protein 2C |
36391 |
0.14 |
| chr10_99051887_99052112 | 0.04 |
ARHGAP19-SLIT1 |
ARHGAP19-SLIT1 readthrough (NMD candidate) |
385 |
0.46 |
| chr17_73511759_73511998 | 0.04 |
CASKIN2 |
CASK interacting protein 2 |
214 |
0.53 |
| chr19_41025424_41025575 | 0.04 |
SPTBN4 |
spectrin, beta, non-erythrocytic 4 |
4952 |
0.16 |
| chr6_33667031_33667259 | 0.04 |
ENSG00000266509 |
. |
1240 |
0.36 |
| chr5_139554573_139554724 | 0.04 |
CYSTM1 |
cysteine-rich transmembrane module containing 1 |
421 |
0.79 |
| chr7_2559962_2560132 | 0.04 |
LFNG |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
551 |
0.71 |
| chr5_172755065_172755216 | 0.04 |
STC2 |
stanniocalcin 2 |
84 |
0.97 |
| chr1_206235900_206236051 | 0.04 |
AVPR1B |
arginine vasopressin receptor 1B |
11999 |
0.16 |
| chr1_57044924_57045096 | 0.04 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
231 |
0.96 |
| chr1_54205166_54205365 | 0.04 |
GLIS1 |
GLIS family zinc finger 1 |
5388 |
0.21 |
| chrX_119149661_119149824 | 0.03 |
RP4-755D9.1 |
|
20459 |
0.16 |
| chr19_13155541_13155731 | 0.03 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
19825 |
0.09 |
| chr15_65823090_65823261 | 0.03 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
83 |
0.96 |
| chr1_227923333_227923484 | 0.03 |
SNAP47 |
synaptosomal-associated protein, 47kDa |
271 |
0.51 |
| chr2_242843726_242843877 | 0.03 |
AC131097.4 |
Protein LOC285095 |
901 |
0.5 |
| chr10_29699092_29699537 | 0.03 |
PTCHD3P1 |
patched domain containing 3 pseudogene 1 |
142 |
0.97 |
| chr17_48637509_48637817 | 0.03 |
CACNA1G |
calcium channel, voltage-dependent, T type, alpha 1G subunit |
766 |
0.34 |
| chr4_40464101_40464252 | 0.03 |
ENSG00000221122 |
. |
1557 |
0.37 |
| chr12_124017770_124017921 | 0.03 |
RILPL1 |
Rab interacting lysosomal protein-like 1 |
420 |
0.78 |
| chr19_51339015_51339203 | 0.03 |
KLK15 |
kallikrein-related peptidase 15 |
1360 |
0.2 |
| chr16_69166636_69166787 | 0.03 |
CIRH1A |
cirrhosis, autosomal recessive 1A (cirhin) |
65 |
0.71 |
| chr12_125011632_125011783 | 0.03 |
NCOR2 |
nuclear receptor corepressor 2 |
8442 |
0.31 |
| chr14_100149110_100149261 | 0.03 |
CYP46A1 |
cytochrome P450, family 46, subfamily A, polypeptide 1 |
1466 |
0.43 |
| chr17_71470730_71470881 | 0.03 |
RP11-449L23.2 |
|
39019 |
0.15 |
| chr2_28616465_28616616 | 0.03 |
FOSL2 |
FOS-like antigen 2 |
815 |
0.43 |
| chr14_65346129_65346280 | 0.03 |
SPTB |
spectrin, beta, erythrocytic |
351 |
0.83 |
| chr11_64878785_64879153 | 0.03 |
AP003068.9 |
|
198 |
0.59 |
| chr1_763102_763253 | 0.03 |
RP11-206L10.8 |
|
17636 |
0.13 |
| chr18_52989024_52989286 | 0.03 |
TCF4 |
transcription factor 4 |
62 |
0.99 |
| chr10_88471165_88471428 | 0.03 |
ENSG00000252189 |
. |
34410 |
0.12 |
| chr12_861875_862026 | 0.03 |
WNK1 |
WNK lysine deficient protein kinase 1 |
139 |
0.97 |
| chr17_40169963_40170444 | 0.03 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
333 |
0.74 |
| chr22_47017266_47017440 | 0.03 |
GRAMD4 |
GRAM domain containing 4 |
1054 |
0.61 |
| chr7_22618226_22618377 | 0.03 |
AC002480.4 |
|
11023 |
0.19 |
| chr1_36929262_36929523 | 0.03 |
MRPS15 |
mitochondrial ribosomal protein S15 |
646 |
0.64 |
| chr17_72856651_72856802 | 0.03 |
GRIN2C |
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
68 |
0.95 |
| chr14_21769078_21769230 | 0.03 |
RPGRIP1 |
retinitis pigmentosa GTPase regulator interacting protein 1 |
13018 |
0.15 |
| chr2_132431451_132431687 | 0.03 |
C2orf27A |
chromosome 2 open reading frame 27A |
48379 |
0.15 |
| chr6_160390514_160390665 | 0.03 |
IGF2R |
insulin-like growth factor 2 receptor |
458 |
0.84 |
| chr13_25592288_25592454 | 0.03 |
LSP1 |
Lymphocyte-specific protein 1; Uncharacterized protein |
830 |
0.67 |
| chr8_146127349_146127646 | 0.03 |
ZNF250 |
zinc finger protein 250 |
56 |
0.97 |
| chr11_119075992_119076532 | 0.03 |
CBL |
Cbl proto-oncogene, E3 ubiquitin protein ligase |
490 |
0.66 |
| chr14_95155916_95156067 | 0.03 |
SERPINA13P |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 13, pseudogene |
48336 |
0.13 |
| chr15_33602917_33603068 | 0.03 |
RP11-489D6.2 |
|
132 |
0.56 |
| chr10_43724787_43724957 | 0.03 |
RASGEF1A |
RasGEF domain family, member 1A |
315 |
0.93 |
| chr14_105554105_105554256 | 0.03 |
GPR132 |
G protein-coupled receptor 132 |
22398 |
0.17 |
| chr7_149558765_149558916 | 0.03 |
RP4-751H13.7 |
|
741 |
0.62 |
| chr17_1532878_1533035 | 0.03 |
SLC43A2 |
solute carrier family 43 (amino acid system L transporter), member 2 |
826 |
0.44 |
| chr10_35104929_35105172 | 0.02 |
PARD3-AS1 |
PARD3 antisense RNA 1 |
355 |
0.75 |
| chr1_33646715_33647017 | 0.02 |
TRIM62 |
tripartite motif containing 62 |
401 |
0.87 |
| chr2_26407379_26407559 | 0.02 |
GAREML |
GRB2 associated, regulator of MAPK1-like |
3884 |
0.23 |
| chr3_195636785_195636941 | 0.02 |
TNK2 |
tyrosine kinase, non-receptor, 2 |
908 |
0.4 |
| chr1_202831073_202831286 | 0.02 |
RP11-480I12.5 |
|
2952 |
0.17 |
| chr2_232479374_232479686 | 0.02 |
C2orf57 |
chromosome 2 open reading frame 57 |
21955 |
0.15 |
| chr9_44751903_44752054 | 0.02 |
ENSG00000238933 |
. |
98243 |
0.08 |
| chr9_97021725_97021876 | 0.02 |
ZNF169 |
zinc finger protein 169 |
190 |
0.94 |
| chr10_93393043_93393253 | 0.02 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
337 |
0.93 |
| chr11_120381906_120382148 | 0.02 |
GRIK4 |
glutamate receptor, ionotropic, kainate 4 |
441 |
0.5 |
| chr4_74904557_74905248 | 0.02 |
CXCL3 |
chemokine (C-X-C motif) ligand 3 |
496 |
0.73 |
| chr2_27071054_27071205 | 0.02 |
DPYSL5 |
dihydropyrimidinase-like 5 |
28 |
0.98 |
| chr19_47951061_47951212 | 0.02 |
SLC8A2 |
solute carrier family 8 (sodium/calcium exchanger), member 2 |
23984 |
0.12 |
| chr4_6909496_6909954 | 0.02 |
TBC1D14 |
TBC1 domain family, member 14 |
1244 |
0.47 |
| chr2_21022702_21022940 | 0.02 |
C2orf43 |
chromosome 2 open reading frame 43 |
9 |
0.99 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.2 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |