Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
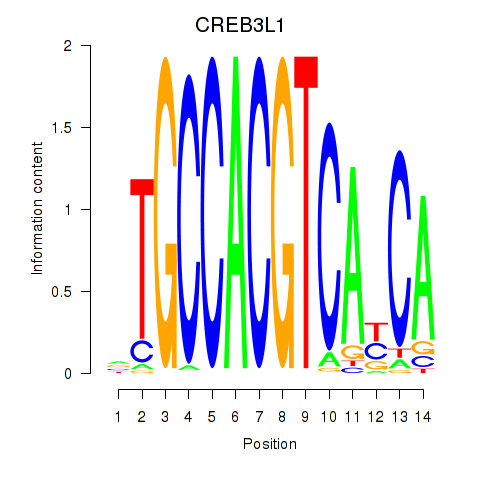
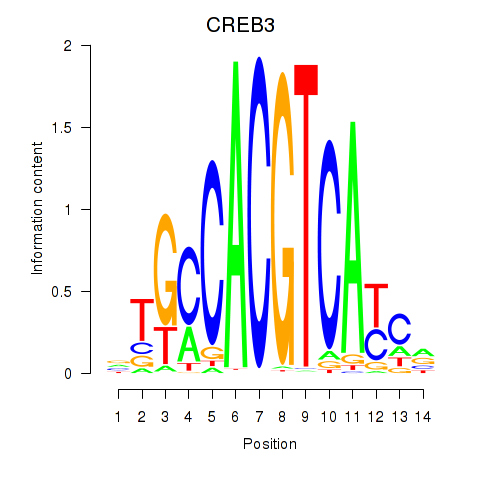
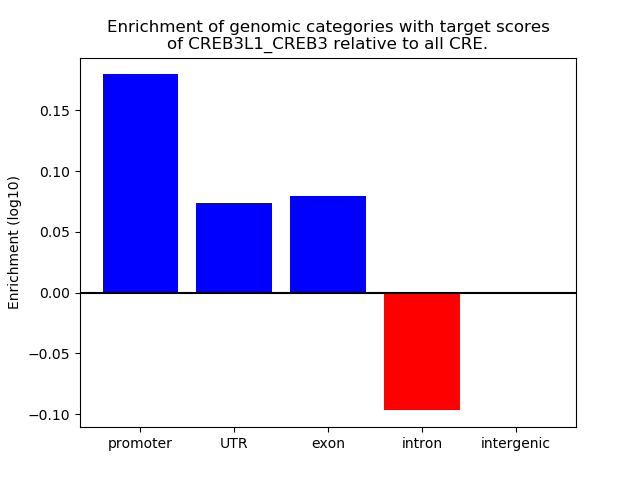
Results for CREB3L1_CREB3
Z-value: 0.86
Transcription factors associated with CREB3L1_CREB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L1
|
ENSG00000157613.6 | cAMP responsive element binding protein 3 like 1 |
|
CREB3
|
ENSG00000107175.6 | cAMP responsive element binding protein 3 |
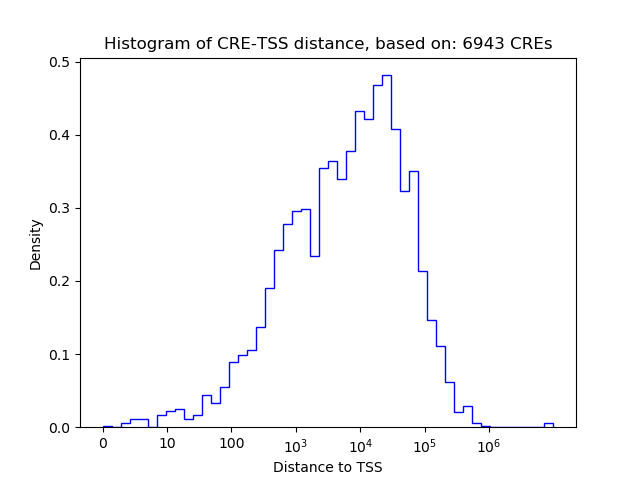
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_35726344_35726495 | CREB3 | 5913 | 0.080709 | -0.59 | 9.8e-02 | Click! |
| chr9_35728507_35728765 | CREB3 | 3696 | 0.092859 | 0.38 | 3.1e-01 | Click! |
| chr9_35731093_35731244 | CREB3 | 1164 | 0.214157 | 0.33 | 3.8e-01 | Click! |
| chr9_35729585_35729939 | CREB3 | 2570 | 0.112031 | 0.31 | 4.1e-01 | Click! |
| chr9_35729999_35730165 | CREB3 | 2250 | 0.122489 | 0.31 | 4.1e-01 | Click! |
| chr11_46300717_46300868 | CREB3L1 | 1564 | 0.362796 | -0.67 | 4.8e-02 | Click! |
| chr11_46315736_46316001 | CREB3L1 | 809 | 0.605315 | -0.64 | 6.3e-02 | Click! |
| chr11_46299587_46299738 | CREB3L1 | 434 | 0.814265 | 0.64 | 6.4e-02 | Click! |
| chr11_46312622_46312792 | CREB3L1 | 3970 | 0.206036 | -0.63 | 6.7e-02 | Click! |
| chr11_46306701_46306852 | CREB3L1 | 7548 | 0.178415 | -0.58 | 1.0e-01 | Click! |
Activity of the CREB3L1_CREB3 motif across conditions
Conditions sorted by the z-value of the CREB3L1_CREB3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
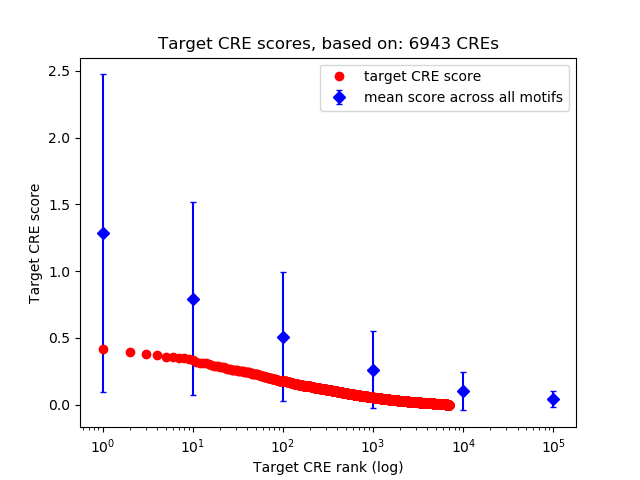
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_179494304_179494455 | 0.42 |
RNF130 |
ring finger protein 130 |
4342 |
0.24 |
| chr21_39776237_39776431 | 0.39 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
94011 |
0.08 |
| chr2_60772484_60772807 | 0.38 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
7895 |
0.25 |
| chr12_110527109_110527260 | 0.37 |
C12orf76 |
chromosome 12 open reading frame 76 |
15693 |
0.19 |
| chr8_133845145_133845296 | 0.36 |
CTC-137K3.1 |
|
6796 |
0.18 |
| chr10_115821373_115821605 | 0.36 |
ADRB1 |
adrenoceptor beta 1 |
17683 |
0.2 |
| chr10_116259271_116259568 | 0.35 |
ABLIM1 |
actin binding LIM protein 1 |
11667 |
0.26 |
| chr3_121804577_121804728 | 0.35 |
CD86 |
CD86 molecule |
5796 |
0.23 |
| chr12_62996382_62996533 | 0.34 |
RP11-631N16.2 |
|
75 |
0.9 |
| chr2_96812616_96812778 | 0.33 |
DUSP2 |
dual specificity phosphatase 2 |
1518 |
0.33 |
| chr20_5654993_5655144 | 0.32 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
63396 |
0.12 |
| chr12_123380878_123381108 | 0.31 |
VPS37B |
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
2 |
0.97 |
| chr6_25171772_25171923 | 0.31 |
ENSG00000222373 |
. |
20794 |
0.18 |
| chr1_59964356_59964507 | 0.31 |
FGGY |
FGGY carbohydrate kinase domain containing |
55159 |
0.17 |
| chr12_114296240_114296499 | 0.30 |
RP11-780K2.1 |
|
74005 |
0.12 |
| chr2_223574012_223574225 | 0.30 |
ENSG00000238852 |
. |
11399 |
0.21 |
| chr6_13294835_13295015 | 0.29 |
RP1-257A7.4 |
|
893 |
0.6 |
| chr6_130009100_130009346 | 0.29 |
ARHGAP18 |
Rho GTPase activating protein 18 |
22147 |
0.24 |
| chr1_160693296_160693447 | 0.29 |
CD48 |
CD48 molecule |
11730 |
0.15 |
| chr8_35729581_35729732 | 0.28 |
AC012215.1 |
Uncharacterized protein |
80291 |
0.12 |
| chr15_98703362_98703675 | 0.28 |
ENSG00000222361 |
. |
85526 |
0.1 |
| chr9_137232344_137232495 | 0.28 |
RXRA |
retinoid X receptor, alpha |
13993 |
0.23 |
| chr6_15090109_15090260 | 0.28 |
ENSG00000242989 |
. |
23015 |
0.23 |
| chr6_135514882_135515033 | 0.27 |
MYB-AS1 |
MYB antisense RNA 1 |
2176 |
0.29 |
| chr20_24823319_24823470 | 0.27 |
CST7 |
cystatin F (leukocystatin) |
106472 |
0.07 |
| chr17_65850368_65850519 | 0.27 |
ENSG00000265086 |
. |
10382 |
0.18 |
| chr17_81041562_81041843 | 0.26 |
METRNL |
meteorin, glial cell differentiation regulator-like |
247 |
0.94 |
| chr3_113326372_113326723 | 0.26 |
ENSG00000265253 |
. |
12824 |
0.16 |
| chr4_80997125_80997368 | 0.26 |
ANTXR2 |
anthrax toxin receptor 2 |
2620 |
0.4 |
| chr7_30188840_30189132 | 0.26 |
MTURN |
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
3473 |
0.2 |
| chr5_36714763_36714914 | 0.26 |
CTD-2353F22.1 |
|
10437 |
0.29 |
| chr5_14332103_14332393 | 0.26 |
TRIO |
trio Rho guanine nucleotide exchange factor |
41162 |
0.22 |
| chr2_73869270_73869546 | 0.25 |
NAT8 |
N-acetyltransferase 8 (GCN5-related, putative) |
112 |
0.97 |
| chr4_144278620_144278790 | 0.25 |
ENSG00000265623 |
. |
14092 |
0.2 |
| chr6_34255201_34255352 | 0.25 |
C6orf1 |
chromosome 6 open reading frame 1 |
38029 |
0.15 |
| chr21_43866253_43866404 | 0.25 |
ENSG00000252619 |
. |
28626 |
0.11 |
| chr19_35939108_35939372 | 0.25 |
FFAR2 |
free fatty acid receptor 2 |
37 |
0.96 |
| chr11_85867980_85868160 | 0.25 |
ENSG00000200877 |
. |
3801 |
0.28 |
| chr5_58858867_58859046 | 0.25 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
23263 |
0.28 |
| chrX_25021375_25021628 | 0.25 |
ARX |
aristaless related homeobox |
12564 |
0.27 |
| chr3_45152072_45152512 | 0.24 |
CDCP1 |
CUB domain containing protein 1 |
35622 |
0.16 |
| chr10_6542814_6542965 | 0.24 |
PRKCQ |
protein kinase C, theta |
79312 |
0.11 |
| chr13_111522529_111522823 | 0.24 |
LINC00346 |
long intergenic non-protein coding RNA 346 |
514 |
0.8 |
| chr2_112368287_112368472 | 0.24 |
ENSG00000266063 |
. |
160332 |
0.04 |
| chr2_31331242_31331393 | 0.23 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
21173 |
0.22 |
| chr11_73181263_73181443 | 0.23 |
RP11-809N8.4 |
|
65011 |
0.09 |
| chr5_178609290_178609441 | 0.23 |
ZNF354C |
zinc finger protein 354C |
121949 |
0.05 |
| chr22_30658397_30658793 | 0.23 |
OSM |
oncostatin M |
3212 |
0.13 |
| chr4_183751040_183751191 | 0.23 |
ENSG00000221186 |
. |
3734 |
0.29 |
| chr19_54703476_54703677 | 0.23 |
AC012314.20 |
|
262 |
0.74 |
| chr6_6970564_6970715 | 0.23 |
ENSG00000240936 |
. |
31509 |
0.21 |
| chr12_111612228_111612379 | 0.23 |
CUX2 |
cut-like homeobox 2 |
75053 |
0.1 |
| chr15_40536086_40536237 | 0.23 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
3315 |
0.14 |
| chr21_39550882_39551033 | 0.22 |
ENSG00000238581 |
. |
8699 |
0.21 |
| chr11_1863672_1864036 | 0.22 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
2430 |
0.15 |
| chr17_79056601_79056878 | 0.22 |
BAIAP2 |
BAI1-associated protein 2 |
3522 |
0.15 |
| chr11_12138065_12138216 | 0.22 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
103 |
0.98 |
| chr9_71362228_71362379 | 0.22 |
PIP5K1B |
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
5132 |
0.26 |
| chr4_38179209_38179752 | 0.22 |
ENSG00000221495 |
. |
60754 |
0.15 |
| chr1_192804077_192804228 | 0.21 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
25981 |
0.21 |
| chrX_14048836_14048989 | 0.21 |
GEMIN8 |
gem (nuclear organelle) associated protein 8 |
900 |
0.68 |
| chr18_77551404_77551663 | 0.21 |
KCNG2 |
potassium voltage-gated channel, subfamily G, member 2 |
72135 |
0.1 |
| chr16_85855178_85855329 | 0.21 |
COX4I1 |
cytochrome c oxidase subunit IV isoform 1 |
20445 |
0.12 |
| chr19_47989123_47989274 | 0.21 |
NAPA-AS1 |
NAPA antisense RNA 1 |
1633 |
0.23 |
| chr10_95196392_95196543 | 0.21 |
MYOF |
myoferlin |
45484 |
0.14 |
| chr17_1528321_1528534 | 0.21 |
SLC43A2 |
solute carrier family 43 (amino acid system L transporter), member 2 |
3208 |
0.14 |
| chr19_440573_440724 | 0.21 |
SHC2 |
SHC (Src homology 2 domain containing) transforming protein 2 |
1618 |
0.25 |
| chr10_75625910_75626399 | 0.20 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
5542 |
0.13 |
| chr10_121410924_121411075 | 0.20 |
BAG3 |
BCL2-associated athanogene 3 |
117 |
0.97 |
| chr1_53102699_53102972 | 0.20 |
FAM159A |
family with sequence similarity 159, member A |
3819 |
0.21 |
| chr8_143815811_143815982 | 0.20 |
CTD-2292P10.2 |
|
6788 |
0.09 |
| chr7_5573568_5573766 | 0.20 |
ACTB |
actin, beta |
3327 |
0.16 |
| chr1_15105044_15105255 | 0.20 |
KAZN |
kazrin, periplakin interacting protein |
145477 |
0.05 |
| chr12_701008_701257 | 0.20 |
RP5-1154L15.1 |
|
1333 |
0.32 |
| chr11_69080924_69081075 | 0.20 |
MYEOV |
myeloma overexpressed |
19374 |
0.26 |
| chr3_178389375_178389526 | 0.20 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
112832 |
0.07 |
| chr2_85762478_85762791 | 0.20 |
ENSG00000266577 |
. |
2885 |
0.13 |
| chr12_132280950_132281101 | 0.20 |
ENSG00000200516 |
. |
4954 |
0.19 |
| chr8_116891196_116891435 | 0.19 |
TRPS1 |
trichorhinophalangeal syndrome I |
69416 |
0.14 |
| chr4_122617356_122617626 | 0.19 |
ANXA5 |
annexin A5 |
626 |
0.78 |
| chr6_6772446_6772597 | 0.19 |
LY86-AS1 |
LY86 antisense RNA 1 |
149517 |
0.04 |
| chr17_78560658_78560826 | 0.19 |
RPTOR |
regulatory associated protein of MTOR, complex 1 |
41474 |
0.17 |
| chr6_42353003_42353575 | 0.19 |
ENSG00000221252 |
. |
26845 |
0.19 |
| chr5_126312490_126312641 | 0.19 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
53935 |
0.15 |
| chr22_46299735_46299886 | 0.19 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
68785 |
0.09 |
| chr20_62493774_62493925 | 0.19 |
C20ORF135 |
|
632 |
0.43 |
| chr17_73860452_73860737 | 0.19 |
WBP2 |
WW domain binding protein 2 |
8006 |
0.08 |
| chr12_68915435_68915586 | 0.18 |
RAP1B |
RAP1B, member of RAS oncogene family |
89109 |
0.08 |
| chr1_28524180_28524503 | 0.18 |
AL353354.2 |
|
2729 |
0.14 |
| chr13_27999670_27999878 | 0.18 |
GTF3A |
general transcription factor IIIA |
1093 |
0.47 |
| chr1_220978316_220978467 | 0.18 |
MARC1 |
mitochondrial amidoxime reducing component 1 |
8294 |
0.19 |
| chr8_6415371_6415522 | 0.18 |
ANGPT2 |
angiopoietin 2 |
5119 |
0.29 |
| chr6_82472333_82472746 | 0.18 |
ENSG00000206886 |
. |
1202 |
0.5 |
| chr19_52270463_52270746 | 0.18 |
FPR2 |
formyl peptide receptor 2 |
1027 |
0.41 |
| chr22_38538827_38538978 | 0.18 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
319 |
0.85 |
| chr16_51375269_51375420 | 0.18 |
SALL1 |
spalt-like transcription factor 1 |
190158 |
0.03 |
| chr3_12945715_12945869 | 0.18 |
AC034198.7 |
|
53634 |
0.1 |
| chrX_130913306_130913457 | 0.18 |
ENSG00000200587 |
. |
159603 |
0.04 |
| chr11_63952754_63952905 | 0.18 |
STIP1 |
stress-induced-phosphoprotein 1 |
85 |
0.93 |
| chr2_62533078_62533236 | 0.18 |
ENSG00000238809 |
. |
41027 |
0.15 |
| chr1_116195861_116196153 | 0.18 |
VANGL1 |
VANGL planar cell polarity protein 1 |
1984 |
0.36 |
| chr7_129347521_129347672 | 0.18 |
ENSG00000212238 |
. |
48510 |
0.11 |
| chr15_78311678_78311855 | 0.18 |
TBC1D2B |
TBC1 domain family, member 2B |
17740 |
0.12 |
| chr1_38156448_38156698 | 0.18 |
C1orf109 |
chromosome 1 open reading frame 109 |
381 |
0.76 |
| chr10_50398424_50398575 | 0.18 |
C10orf128 |
chromosome 10 open reading frame 128 |
2063 |
0.33 |
| chr16_72942017_72942168 | 0.18 |
ENSG00000221799 |
. |
76664 |
0.08 |
| chr21_40362776_40362927 | 0.17 |
ENSG00000272015 |
. |
96142 |
0.08 |
| chr2_112763039_112763349 | 0.17 |
MERTK |
c-mer proto-oncogene tyrosine kinase |
13796 |
0.22 |
| chr17_80833585_80833738 | 0.17 |
TBCD |
tubulin folding cofactor D |
8367 |
0.18 |
| chr9_101984883_101985113 | 0.17 |
SEC61B |
Sec61 beta subunit |
415 |
0.66 |
| chr2_47839910_47840061 | 0.17 |
AC138655.1 |
CDNA: FLJ23120 fis, clone LNG07989; HCG1987724; Uncharacterized protein |
40384 |
0.14 |
| chr20_7997297_7997448 | 0.17 |
RP5-971N18.3 |
|
2498 |
0.3 |
| chr20_61555156_61555835 | 0.17 |
DIDO1 |
death inducer-obliterator 1 |
2326 |
0.23 |
| chr11_119560847_119560998 | 0.17 |
ENSG00000199217 |
. |
33901 |
0.14 |
| chr11_44938433_44938696 | 0.17 |
TSPAN18 |
tetraspanin 18 |
10606 |
0.25 |
| chr22_35933430_35933581 | 0.17 |
RASD2 |
RASD family, member 2 |
3410 |
0.3 |
| chr19_45612723_45613017 | 0.17 |
PPP1R37 |
protein phosphatase 1, regulatory subunit 37 |
16438 |
0.08 |
| chr1_1101221_1101488 | 0.17 |
ENSG00000207730 |
. |
1130 |
0.25 |
| chr21_43953040_43953239 | 0.17 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
18505 |
0.13 |
| chr12_121665055_121665206 | 0.17 |
P2RX4 |
purinergic receptor P2X, ligand-gated ion channel, 4 |
17162 |
0.17 |
| chrX_65191097_65191248 | 0.17 |
RP6-159A1.3 |
|
28421 |
0.2 |
| chr16_75145410_75145702 | 0.17 |
RP11-252E2.1 |
|
946 |
0.49 |
| chr10_49907092_49907243 | 0.16 |
WDFY4 |
WDFY family member 4 |
10584 |
0.23 |
| chr1_159906503_159906698 | 0.16 |
IGSF9 |
immunoglobulin superfamily, member 9 |
8786 |
0.1 |
| chr17_47590468_47590727 | 0.16 |
NGFR |
nerve growth factor receptor |
15844 |
0.14 |
| chr19_13215621_13215858 | 0.16 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
1764 |
0.21 |
| chr6_26181116_26181267 | 0.16 |
HIST1H2BE |
histone cluster 1, H2be |
2767 |
0.09 |
| chr19_45253255_45253406 | 0.16 |
BCL3 |
B-cell CLL/lymphoma 3 |
1248 |
0.31 |
| chr5_169855198_169855349 | 0.16 |
KCNMB1 |
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
38592 |
0.14 |
| chr16_19124977_19125163 | 0.16 |
ITPRIPL2 |
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
184 |
0.93 |
| chr2_112251056_112251269 | 0.16 |
ENSG00000266139 |
. |
172494 |
0.03 |
| chr7_1859118_1859366 | 0.16 |
AC110781.3 |
Protein LOC100128374 |
18980 |
0.17 |
| chr3_28389894_28390045 | 0.16 |
AZI2 |
5-azacytidine induced 2 |
1 |
0.94 |
| chr14_91252167_91252318 | 0.16 |
TTC7B |
tetratricopeptide repeat domain 7B |
339 |
0.92 |
| chr10_5567264_5567415 | 0.16 |
RP11-116G8.5 |
|
366 |
0.46 |
| chr7_47616572_47616723 | 0.16 |
TNS3 |
tensin 3 |
4582 |
0.32 |
| chr9_34170162_34170313 | 0.16 |
UBAP1 |
ubiquitin associated protein 1 |
8766 |
0.15 |
| chr19_57683108_57683259 | 0.16 |
DUXA |
double homeobox A |
4372 |
0.15 |
| chr2_89157495_89157741 | 0.16 |
IGKJ5 |
immunoglobulin kappa joining 5 |
2499 |
0.12 |
| chr17_47486265_47486624 | 0.16 |
RP11-1079K10.4 |
|
5001 |
0.13 |
| chr11_129695943_129696094 | 0.16 |
TMEM45B |
transmembrane protein 45B |
10304 |
0.27 |
| chr17_35658363_35658514 | 0.16 |
ACACA |
acetyl-CoA carboxylase alpha |
1746 |
0.42 |
| chr2_60595552_60595831 | 0.16 |
ENSG00000200807 |
. |
16049 |
0.22 |
| chr22_23909098_23909249 | 0.16 |
AP000345.2 |
|
78 |
0.97 |
| chr4_37853152_37853303 | 0.16 |
PGM2 |
phosphoglucomutase 2 |
24884 |
0.21 |
| chr1_162319832_162320115 | 0.15 |
ENSG00000207729 |
. |
7637 |
0.11 |
| chr12_7905511_7905662 | 0.15 |
CLEC4C |
C-type lectin domain family 4, member C |
1385 |
0.3 |
| chr15_68502161_68502336 | 0.15 |
CALML4 |
calmodulin-like 4 |
3831 |
0.2 |
| chr20_31256682_31256833 | 0.15 |
RP11-410N8.4 |
|
67318 |
0.08 |
| chr19_16552842_16553129 | 0.15 |
CTD-2013N17.4 |
|
1228 |
0.36 |
| chr1_27560545_27560765 | 0.15 |
RP11-40H20.4 |
|
188 |
0.68 |
| chr16_646360_646511 | 0.15 |
RAB40C |
RAB40C, member RAS oncogene family |
6103 |
0.07 |
| chr12_58128131_58128282 | 0.15 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
2594 |
0.1 |
| chr11_121454430_121454581 | 0.15 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
6623 |
0.32 |
| chr11_11523784_11523935 | 0.15 |
RP11-483L5.1 |
|
67772 |
0.12 |
| chr16_87802997_87803148 | 0.15 |
KLHDC4 |
kelch domain containing 4 |
3490 |
0.22 |
| chr22_44464688_44464966 | 0.15 |
PARVB |
parvin, beta |
114 |
0.98 |
| chr2_43633250_43633401 | 0.15 |
ENSG00000252804 |
. |
2121 |
0.44 |
| chr2_238634076_238634227 | 0.15 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
33169 |
0.17 |
| chr12_7031787_7032018 | 0.15 |
ENO2 |
enolase 2 (gamma, neuronal) |
1158 |
0.2 |
| chr1_204332619_204332786 | 0.15 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
3658 |
0.22 |
| chr5_123646330_123646481 | 0.15 |
RP11-436H11.2 |
|
418119 |
0.01 |
| chr11_60914868_60915019 | 0.15 |
VPS37C |
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
13665 |
0.16 |
| chr12_122230072_122230277 | 0.14 |
RHOF |
ras homolog family member F (in filopodia) |
1092 |
0.46 |
| chr7_129007754_129007992 | 0.14 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
91 |
0.97 |
| chr16_66606248_66606582 | 0.14 |
CMTM1 |
CKLF-like MARVEL transmembrane domain containing 1 |
5998 |
0.1 |
| chr5_58055121_58055272 | 0.14 |
RP11-479O16.1 |
|
25410 |
0.24 |
| chr12_107350540_107350837 | 0.14 |
C12orf23 |
chromosome 12 open reading frame 23 |
125 |
0.94 |
| chr15_67833013_67833278 | 0.14 |
MAP2K5 |
mitogen-activated protein kinase kinase 5 |
1902 |
0.39 |
| chr5_130868870_130869280 | 0.14 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
349 |
0.93 |
| chr18_29210689_29211071 | 0.14 |
ENSG00000252379 |
. |
38461 |
0.11 |
| chr19_28964296_28964459 | 0.14 |
ENSG00000238514 |
. |
457533 |
0.01 |
| chr19_6059787_6060020 | 0.14 |
RFX2 |
regulatory factor X, 2 (influences HLA class II expression) |
2609 |
0.21 |
| chr14_86528076_86528227 | 0.14 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
531579 |
0.0 |
| chr10_101310900_101311051 | 0.14 |
NKX2-3 |
NK2 homeobox 3 |
18285 |
0.15 |
| chr22_43581545_43581696 | 0.14 |
TTLL12 |
tubulin tyrosine ligase-like family, member 12 |
1519 |
0.32 |
| chr3_176888184_176888459 | 0.14 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
25890 |
0.2 |
| chr1_25307295_25307446 | 0.14 |
RUNX3 |
runt-related transcription factor 3 |
15869 |
0.19 |
| chr3_107837151_107837302 | 0.14 |
CD47 |
CD47 molecule |
27354 |
0.24 |
| chr9_95523817_95524060 | 0.14 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
3156 |
0.26 |
| chr19_11159757_11160063 | 0.14 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
9601 |
0.14 |
| chr12_11897037_11897481 | 0.14 |
ETV6 |
ets variant 6 |
8176 |
0.3 |
| chr21_35709927_35710173 | 0.14 |
KCNE2 |
potassium voltage-gated channel, Isk-related family, member 2 |
26273 |
0.13 |
| chr5_171059986_171060137 | 0.14 |
ENSG00000264303 |
. |
137473 |
0.04 |
| chr2_73263307_73263458 | 0.14 |
SFXN5 |
sideroflexin 5 |
35420 |
0.16 |
| chr22_28037496_28037647 | 0.14 |
RP11-375H17.1 |
|
74897 |
0.12 |
| chr2_173850166_173850396 | 0.14 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
41073 |
0.18 |
| chr13_99566346_99566616 | 0.14 |
DOCK9 |
dedicator of cytokinesis 9 |
58243 |
0.13 |
| chr3_128711977_128712128 | 0.14 |
KIAA1257 |
KIAA1257 |
867 |
0.53 |
| chr15_32948468_32948619 | 0.14 |
SCG5 |
secretogranin V (7B2 protein) |
12719 |
0.14 |
| chr2_3583541_3583693 | 0.14 |
RP13-512J5.1 |
Uncharacterized protein |
11930 |
0.12 |
| chr11_4489038_4489189 | 0.14 |
OR52K2 |
olfactory receptor, family 52, subfamily K, member 2 |
18588 |
0.13 |
| chr10_119303115_119303318 | 0.14 |
EMX2 |
empty spiracles homeobox 2 |
707 |
0.63 |
| chr3_142720005_142720156 | 0.14 |
U2SURP |
U2 snRNP-associated SURP domain containing |
286 |
0.91 |
| chr20_30184012_30184163 | 0.14 |
ID1 |
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
8999 |
0.11 |
| chr19_52552377_52552528 | 0.14 |
ZNF432 |
zinc finger protein 432 |
379 |
0.77 |
| chr20_60719765_60720239 | 0.14 |
SS18L1 |
synovial sarcoma translocation gene on chromosome 18-like 1 |
1151 |
0.34 |
| chr15_89165477_89165628 | 0.14 |
AEN |
apoptosis enhancing nuclease |
951 |
0.49 |
| chr17_80308630_80308781 | 0.14 |
TEX19 |
testis expressed 19 |
8418 |
0.1 |
| chr1_20905252_20905403 | 0.14 |
CDA |
cytidine deaminase |
10114 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0021894 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0003079 | obsolete positive regulation of natriuresis(GO:0003079) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.0 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.3 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.0 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0060013 | righting reflex(GO:0060013) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0032356 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |