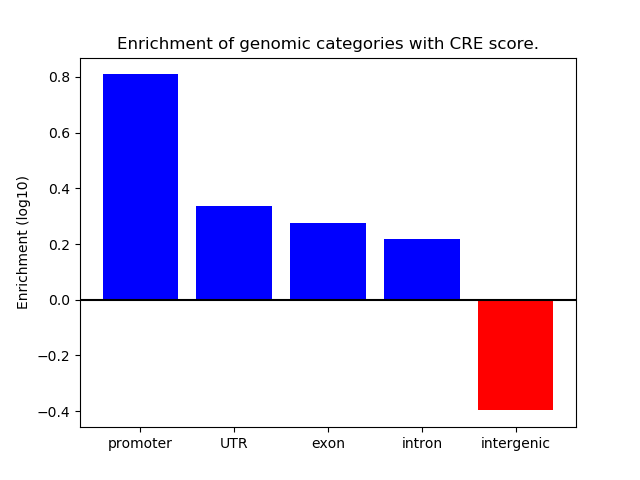
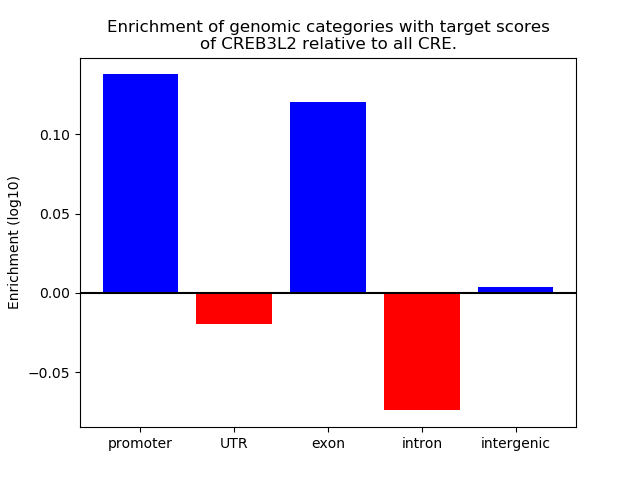
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
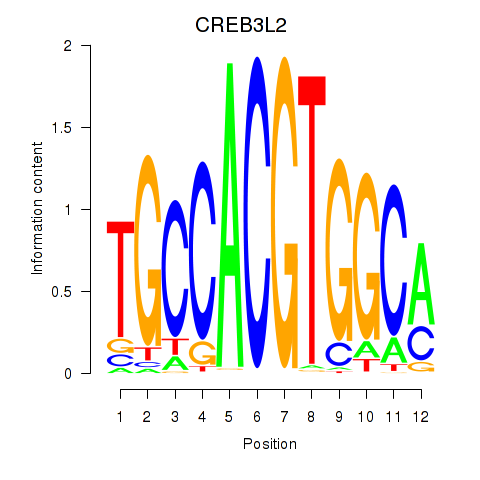
Results for CREB3L2
Z-value: 1.06
Transcription factors associated with CREB3L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L2
|
ENSG00000182158.10 | cAMP responsive element binding protein 3 like 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_137668727_137668878 | CREB3L2 | 17991 | 0.183352 | 0.78 | 1.3e-02 | Click! |
| chr7_137663575_137663726 | CREB3L2 | 23143 | 0.172034 | 0.77 | 1.5e-02 | Click! |
| chr7_137573299_137573450 | CREB3L2 | 39641 | 0.153479 | 0.77 | 1.6e-02 | Click! |
| chr7_137605978_137606391 | CREB3L2 | 6831 | 0.230519 | -0.77 | 1.6e-02 | Click! |
| chr7_137621255_137621573 | CREB3L2 | 2778 | 0.292325 | 0.74 | 2.2e-02 | Click! |
Activity of the CREB3L2 motif across conditions
Conditions sorted by the z-value of the CREB3L2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
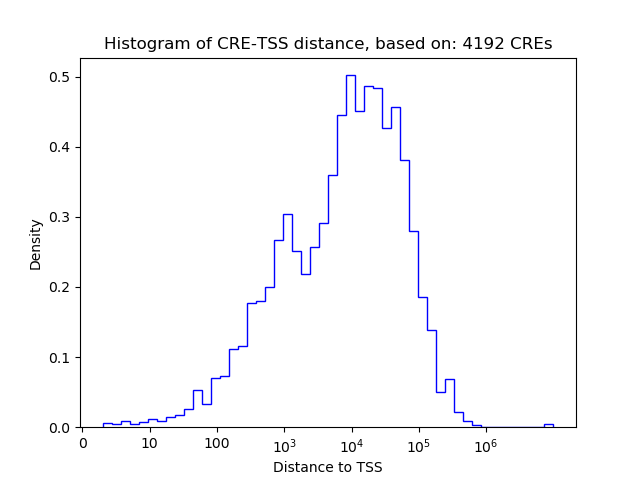
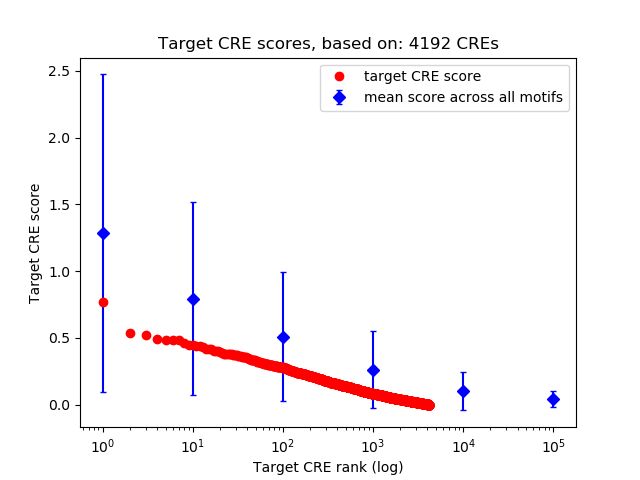
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_16681395_16681795 | 0.77 |
RP1-151F17.1 |
|
79774 |
0.1 |
| chr17_48718114_48718630 | 0.54 |
ABCC3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
6154 |
0.13 |
| chr14_100906069_100906661 | 0.52 |
RP11-362L22.1 |
|
33123 |
0.1 |
| chr15_68683192_68683351 | 0.49 |
ITGA11 |
integrin, alpha 11 |
41221 |
0.18 |
| chr6_147235336_147235538 | 0.49 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
1584 |
0.55 |
| chr16_29167036_29167187 | 0.49 |
CTB-134H23.3 |
|
48345 |
0.11 |
| chr8_126187977_126188128 | 0.49 |
NSMCE2 |
non-SMC element 2, MMS21 homolog (S. cerevisiae) |
51666 |
0.14 |
| chr4_129308818_129308969 | 0.46 |
PGRMC2 |
progesterone receptor membrane component 2 |
98909 |
0.09 |
| chr9_137394449_137394849 | 0.44 |
RXRA |
retinoid X receptor, alpha |
96221 |
0.07 |
| chr18_53398894_53399045 | 0.44 |
TCF4 |
transcription factor 4 |
66951 |
0.14 |
| chr2_223574012_223574225 | 0.44 |
ENSG00000238852 |
. |
11399 |
0.21 |
| chr14_91281521_91281794 | 0.44 |
TTC7B |
tetratricopeptide repeat domain 7B |
1104 |
0.63 |
| chr1_209831123_209831274 | 0.43 |
LAMB3 |
laminin, beta 3 |
5387 |
0.15 |
| chr5_118733387_118734036 | 0.42 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
41981 |
0.13 |
| chr10_1290240_1290555 | 0.42 |
ADARB2 |
adenosine deaminase, RNA-specific, B2 (non-functional) |
44072 |
0.17 |
| chr5_71475626_71475777 | 0.42 |
MAP1B |
microtubule-associated protein 1B |
246 |
0.94 |
| chr21_35709927_35710173 | 0.41 |
KCNE2 |
potassium voltage-gated channel, Isk-related family, member 2 |
26273 |
0.13 |
| chr7_141615794_141616062 | 0.40 |
OR9A4 |
olfactory receptor, family 9, subfamily A, member 4 |
2689 |
0.16 |
| chr2_71682494_71682648 | 0.40 |
DYSF |
dysferlin |
1719 |
0.44 |
| chr4_120986498_120987561 | 0.39 |
RP11-679C8.2 |
|
1084 |
0.42 |
| chr22_42765026_42765177 | 0.38 |
TCF20 |
transcription factor 20 (AR1) |
25479 |
0.18 |
| chr9_94660876_94661276 | 0.38 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
50086 |
0.18 |
| chr21_30754344_30754495 | 0.38 |
ENSG00000239171 |
. |
7391 |
0.18 |
| chr6_42283087_42283238 | 0.38 |
ENSG00000221252 |
. |
96972 |
0.06 |
| chr10_79638365_79638516 | 0.38 |
AL391421.1 |
Uncharacterized protein; cDNA FLJ43696 fis, clone TBAES2007964 |
11807 |
0.18 |
| chr3_139020939_139021090 | 0.38 |
MRPS22 |
mitochondrial ribosomal protein S22 |
41847 |
0.13 |
| chr1_201237669_201237867 | 0.38 |
PKP1 |
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) |
14812 |
0.18 |
| chr1_145526041_145526192 | 0.37 |
ITGA10 |
integrin, alpha 10 |
1081 |
0.31 |
| chr1_8882630_8882823 | 0.37 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
5024 |
0.19 |
| chr16_11295208_11295453 | 0.37 |
RMI2 |
RecQ mediated genome instability 2 |
48176 |
0.08 |
| chr15_77713627_77714545 | 0.37 |
HMG20A |
high mobility group 20A |
733 |
0.64 |
| chr5_134773694_134773956 | 0.36 |
C5orf20 |
chromosome 5 open reading frame 20 |
9213 |
0.14 |
| chr11_67896240_67896572 | 0.36 |
CTD-2655K5.1 |
|
859 |
0.55 |
| chr3_29376763_29376914 | 0.36 |
ENSG00000216169 |
. |
34074 |
0.18 |
| chr18_22878825_22879093 | 0.36 |
CTD-2006O16.2 |
|
3131 |
0.37 |
| chr6_52379346_52379497 | 0.36 |
TRAM2 |
translocation associated membrane protein 2 |
62292 |
0.12 |
| chr15_35598915_35599148 | 0.36 |
ENSG00000265102 |
. |
65534 |
0.14 |
| chr2_112763039_112763349 | 0.35 |
MERTK |
c-mer proto-oncogene tyrosine kinase |
13796 |
0.22 |
| chr20_25208276_25208481 | 0.35 |
AL035252.1 |
HCG2018895; Uncharacterized protein |
1008 |
0.49 |
| chr1_33849901_33850220 | 0.35 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
8866 |
0.16 |
| chr1_59602387_59602647 | 0.35 |
FGGY |
FGGY carbohydrate kinase domain containing |
159793 |
0.04 |
| chr6_145569007_145569263 | 0.34 |
ENSG00000221796 |
. |
100759 |
0.09 |
| chr10_134330476_134330668 | 0.34 |
LINC01165 |
long intergenic non-protein coding RNA 1165 |
5381 |
0.22 |
| chr1_41467777_41468071 | 0.34 |
CTPS1 |
CTP synthase 1 |
19091 |
0.19 |
| chr21_39660692_39660843 | 0.33 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
7712 |
0.29 |
| chr16_72405306_72405606 | 0.33 |
ENSG00000207514 |
. |
5147 |
0.31 |
| chr12_48147567_48147718 | 0.33 |
SLC48A1 |
solute carrier family 48 (heme transporter), member 1 |
57 |
0.96 |
| chr16_55530572_55531230 | 0.33 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
8327 |
0.22 |
| chr6_13397775_13398015 | 0.33 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
10474 |
0.21 |
| chr14_61789232_61789447 | 0.33 |
PRKCH |
protein kinase C, eta |
43 |
0.71 |
| chr15_90325381_90325563 | 0.32 |
MESP2 |
mesoderm posterior 2 homolog (mouse) |
5883 |
0.14 |
| chr6_82575347_82575498 | 0.32 |
ENSG00000206886 |
. |
101681 |
0.07 |
| chr4_57945352_57945503 | 0.32 |
ENSG00000238541 |
. |
27084 |
0.14 |
| chr18_56658261_56658433 | 0.32 |
ENSG00000251870 |
. |
64819 |
0.11 |
| chr1_107480050_107480201 | 0.32 |
PRMT6 |
protein arginine methyltransferase 6 |
119142 |
0.07 |
| chr1_37920990_37921188 | 0.32 |
LINC01137 |
long intergenic non-protein coding RNA 1137 |
18923 |
0.15 |
| chr2_112375416_112375598 | 0.31 |
ENSG00000266063 |
. |
153204 |
0.04 |
| chr12_124870235_124870636 | 0.31 |
NCOR2 |
nuclear receptor corepressor 2 |
2935 |
0.37 |
| chr10_115821373_115821605 | 0.31 |
ADRB1 |
adrenoceptor beta 1 |
17683 |
0.2 |
| chr1_33798502_33798653 | 0.31 |
ENSG00000266239 |
. |
484 |
0.7 |
| chr10_79291591_79291742 | 0.31 |
ENSG00000199592 |
. |
55141 |
0.15 |
| chr5_159698543_159698722 | 0.31 |
ENSG00000243654 |
. |
20163 |
0.16 |
| chr16_85424724_85424875 | 0.31 |
RP11-680G10.1 |
Uncharacterized protein |
33730 |
0.18 |
| chr3_122785668_122785819 | 0.31 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
174 |
0.96 |
| chr9_124051158_124051423 | 0.31 |
GSN |
gelsolin |
2420 |
0.21 |
| chr10_102124571_102124984 | 0.31 |
SCD |
stearoyl-CoA desaturase (delta-9-desaturase) |
17896 |
0.13 |
| chr9_6973383_6973534 | 0.30 |
KDM4C |
lysine (K)-specific demethylase 4C |
40330 |
0.2 |
| chr11_69402239_69402390 | 0.30 |
CCND1 |
cyclin D1 |
53541 |
0.14 |
| chr7_47616572_47616723 | 0.30 |
TNS3 |
tensin 3 |
4582 |
0.32 |
| chr6_7921829_7921980 | 0.30 |
TXNDC5 |
thioredoxin domain containing 5 (endoplasmic reticulum) |
10857 |
0.3 |
| chr1_234688974_234689202 | 0.30 |
ENSG00000212144 |
. |
39933 |
0.14 |
| chr7_1595634_1595900 | 0.30 |
TMEM184A |
transmembrane protein 184A |
11 |
0.97 |
| chr20_5844973_5845124 | 0.30 |
ENSG00000202380 |
. |
25653 |
0.17 |
| chr1_45793013_45793296 | 0.30 |
HPDL |
4-hydroxyphenylpyruvate dioxygenase-like |
609 |
0.67 |
| chr13_20764967_20765118 | 0.30 |
GJB2 |
gap junction protein, beta 2, 26kDa |
1123 |
0.49 |
| chr17_75307827_75307996 | 0.29 |
SEPT9 |
septin 9 |
7686 |
0.23 |
| chr1_15105044_15105255 | 0.29 |
KAZN |
kazrin, periplakin interacting protein |
145477 |
0.05 |
| chr19_18959817_18960274 | 0.29 |
UPF1 |
UPF1 regulator of nonsense transcripts homolog (yeast) |
17235 |
0.11 |
| chr2_151337576_151337727 | 0.29 |
RND3 |
Rho family GTPase 3 |
4245 |
0.38 |
| chr16_700261_700412 | 0.29 |
WDR90 |
WD repeat domain 90 |
919 |
0.22 |
| chr17_75346852_75347029 | 0.29 |
SEPT9 |
septin 9 |
22332 |
0.16 |
| chr1_27916174_27916325 | 0.29 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
13853 |
0.15 |
| chr9_127030805_127030956 | 0.29 |
RP11-121A14.3 |
|
5725 |
0.18 |
| chr4_69331756_69331907 | 0.29 |
TMPRSS11E |
transmembrane protease, serine 11E |
18664 |
0.25 |
| chr8_18696899_18697278 | 0.29 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
14796 |
0.23 |
| chr1_25040508_25040659 | 0.29 |
CLIC4 |
chloride intracellular channel 4 |
31265 |
0.15 |
| chr12_14413818_14414239 | 0.29 |
ENSG00000251908 |
. |
10753 |
0.22 |
| chr21_40144936_40145370 | 0.29 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
32078 |
0.22 |
| chr20_30195413_30195568 | 0.29 |
ENSG00000264395 |
. |
501 |
0.67 |
| chr22_37958323_37958474 | 0.29 |
CDC42EP1 |
CDC42 effector protein (Rho GTPase binding) 1 |
1263 |
0.33 |
| chr11_57282126_57282281 | 0.28 |
SLC43A1 |
solute carrier family 43 (amino acid system L transporter), member 1 |
115 |
0.93 |
| chr5_114795818_114795969 | 0.28 |
FEM1C |
fem-1 homolog c (C. elegans) |
84698 |
0.08 |
| chr13_98953540_98953691 | 0.28 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
30397 |
0.16 |
| chr16_3900112_3900331 | 0.28 |
CREBBP |
CREB binding protein |
29900 |
0.18 |
| chr9_91390340_91390723 | 0.28 |
ENSG00000265873 |
. |
29711 |
0.24 |
| chr10_77471952_77472136 | 0.28 |
RP11-367B6.2 |
|
30454 |
0.19 |
| chr1_9127158_9127594 | 0.28 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
2228 |
0.26 |
| chr8_23396820_23397037 | 0.28 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
10355 |
0.16 |
| chr17_35534355_35534506 | 0.28 |
ACACA |
acetyl-CoA carboxylase alpha |
10804 |
0.24 |
| chr5_14153391_14153542 | 0.28 |
TRIO |
trio Rho guanine nucleotide exchange factor |
9637 |
0.32 |
| chr16_57411779_57412024 | 0.28 |
CX3CL1 |
chemokine (C-X3-C motif) ligand 1 |
1017 |
0.45 |
| chr7_101688884_101689543 | 0.28 |
CTA-357J21.1 |
|
12910 |
0.24 |
| chr9_138818669_138818820 | 0.28 |
RP11-432J22.2 |
|
19086 |
0.17 |
| chr19_35939108_35939372 | 0.28 |
FFAR2 |
free fatty acid receptor 2 |
37 |
0.96 |
| chr6_83075536_83075687 | 0.27 |
TPBG |
trophoblast glycoprotein |
1650 |
0.53 |
| chr1_62070273_62070424 | 0.27 |
ENSG00000264551 |
. |
24355 |
0.25 |
| chrX_52964621_52965041 | 0.27 |
FAM156A |
family with sequence similarity 156, member A |
20698 |
0.18 |
| chr9_109622867_109623098 | 0.27 |
ZNF462 |
zinc finger protein 462 |
2396 |
0.34 |
| chr18_59658543_59658754 | 0.27 |
RNF152 |
ring finger protein 152 |
97184 |
0.08 |
| chr17_76127130_76127305 | 0.27 |
TMC8 |
transmembrane channel-like 8 |
348 |
0.54 |
| chr10_81160624_81160775 | 0.27 |
RP11-342M3.5 |
|
18615 |
0.19 |
| chr13_50772718_50772869 | 0.27 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
116486 |
0.05 |
| chr6_170501787_170501938 | 0.27 |
RP11-302L19.1 |
|
24121 |
0.21 |
| chr6_26296598_26296749 | 0.27 |
HIST1H4H |
histone cluster 1, H4h |
10911 |
0.08 |
| chr22_43411349_43411733 | 0.26 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
390 |
0.86 |
| chr15_70147676_70147827 | 0.26 |
ENSG00000215958 |
. |
69908 |
0.12 |
| chr17_73937351_73937691 | 0.26 |
FBF1 |
Fas (TNFRSF6) binding factor 1 |
402 |
0.69 |
| chr16_839632_839783 | 0.26 |
CHTF18 |
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae) |
394 |
0.62 |
| chr9_137366473_137366866 | 0.26 |
RXRA |
retinoid X receptor, alpha |
68241 |
0.11 |
| chr14_56617754_56618009 | 0.26 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
32054 |
0.21 |
| chr14_77773813_77773964 | 0.26 |
POMT2 |
protein-O-mannosyltransferase 2 |
12795 |
0.12 |
| chr8_122360906_122361134 | 0.26 |
ENSG00000221644 |
. |
161888 |
0.04 |
| chr9_137250637_137250831 | 0.26 |
ENSG00000263897 |
. |
20523 |
0.22 |
| chr15_82180102_82180253 | 0.26 |
ENSG00000222521 |
. |
156458 |
0.04 |
| chr10_119303115_119303318 | 0.26 |
EMX2 |
empty spiracles homeobox 2 |
707 |
0.63 |
| chr6_151244201_151244352 | 0.25 |
ENSG00000202119 |
. |
16082 |
0.21 |
| chr1_199403544_199403695 | 0.25 |
ENSG00000263805 |
. |
180789 |
0.03 |
| chr19_50804315_50804466 | 0.25 |
CTB-191K22.5 |
|
9406 |
0.12 |
| chr5_74867541_74867832 | 0.25 |
POLK |
polymerase (DNA directed) kappa |
24838 |
0.14 |
| chr8_142143433_142143584 | 0.25 |
RP11-809O17.1 |
|
3448 |
0.24 |
| chr6_143388789_143389162 | 0.25 |
AIG1 |
androgen-induced 1 |
6937 |
0.19 |
| chr14_104018069_104018220 | 0.25 |
ENSG00000252469 |
. |
1462 |
0.17 |
| chr19_5038576_5038872 | 0.25 |
KDM4B |
lysine (K)-specific demethylase 4B |
22388 |
0.2 |
| chr6_15090109_15090260 | 0.25 |
ENSG00000242989 |
. |
23015 |
0.23 |
| chr11_126455523_126455674 | 0.25 |
KIRREL3-AS1 |
KIRREL3 antisense RNA 1 |
41756 |
0.17 |
| chr14_75687131_75687282 | 0.25 |
ENSG00000252013 |
. |
18334 |
0.14 |
| chr3_142840553_142840704 | 0.25 |
CHST2 |
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
2455 |
0.34 |
| chr17_72837180_72837562 | 0.25 |
GRIN2C |
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
18627 |
0.11 |
| chr15_63526608_63526933 | 0.25 |
APH1B |
APH1B gamma secretase subunit |
41447 |
0.14 |
| chr8_56851680_56851967 | 0.24 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
970 |
0.5 |
| chr20_33301034_33301185 | 0.24 |
TP53INP2 |
tumor protein p53 inducible nuclear protein 2 |
8570 |
0.21 |
| chr15_43545776_43545927 | 0.24 |
ENSG00000202211 |
. |
5973 |
0.13 |
| chr7_121170101_121170252 | 0.24 |
ENSG00000221690 |
. |
44632 |
0.19 |
| chr1_86035839_86035990 | 0.24 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
8019 |
0.19 |
| chr22_46501298_46501449 | 0.24 |
ENSG00000198986 |
. |
7256 |
0.1 |
| chr17_1459503_1459654 | 0.24 |
PITPNA |
phosphatidylinositol transfer protein, alpha |
3212 |
0.17 |
| chr1_60598722_60598873 | 0.24 |
C1orf87 |
chromosome 1 open reading frame 87 |
59355 |
0.16 |
| chr3_58037044_58037226 | 0.24 |
FLNB |
filamin B, beta |
26921 |
0.22 |
| chr7_372645_372959 | 0.24 |
AC187652.1 |
Protein LOC100996433 |
69884 |
0.12 |
| chr1_156680056_156680253 | 0.24 |
RP11-66D17.5 |
|
2812 |
0.13 |
| chr5_1515989_1516747 | 0.24 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
7724 |
0.23 |
| chr1_235104740_235105149 | 0.24 |
ENSG00000239690 |
. |
65011 |
0.12 |
| chr19_7897788_7897939 | 0.24 |
EVI5L |
ecotropic viral integration site 5-like |
2744 |
0.16 |
| chr12_46782989_46783140 | 0.24 |
SLC38A2 |
solute carrier family 38, member 2 |
16414 |
0.23 |
| chr11_72295622_72295773 | 0.24 |
RP11-169D4.2 |
|
81 |
0.64 |
| chr13_110417364_110417521 | 0.24 |
IRS2 |
insulin receptor substrate 2 |
21473 |
0.23 |
| chr14_78266699_78267170 | 0.24 |
ADCK1 |
aarF domain containing kinase 1 |
452 |
0.79 |
| chr18_60118168_60118548 | 0.24 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
71882 |
0.1 |
| chr19_632822_632973 | 0.23 |
POLRMT |
polymerase (RNA) mitochondrial (DNA directed) |
700 |
0.47 |
| chr17_29896770_29896921 | 0.23 |
AC003101.1 |
Uncharacterized protein |
1316 |
0.29 |
| chr3_71591005_71591175 | 0.23 |
ENSG00000221264 |
. |
150 |
0.95 |
| chr4_5748628_5748779 | 0.23 |
EVC |
Ellis van Creveld syndrome |
35761 |
0.18 |
| chr1_246599104_246599255 | 0.23 |
ENSG00000207326 |
. |
9758 |
0.22 |
| chr3_178389375_178389526 | 0.23 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
112832 |
0.07 |
| chr1_214668072_214668223 | 0.23 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
30001 |
0.23 |
| chr8_80681240_80681519 | 0.23 |
HEY1 |
hes-related family bHLH transcription factor with YRPW motif 1 |
1281 |
0.55 |
| chr19_19488516_19488930 | 0.23 |
GATAD2A |
GATA zinc finger domain containing 2A |
7912 |
0.14 |
| chr12_109080507_109080751 | 0.23 |
CORO1C |
coronin, actin binding protein, 1C |
9549 |
0.15 |
| chr13_32059321_32059637 | 0.23 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
254195 |
0.02 |
| chr9_90094686_90094877 | 0.23 |
DAPK1 |
death-associated protein kinase 1 |
17362 |
0.26 |
| chr6_6916729_6917061 | 0.23 |
ENSG00000240936 |
. |
22235 |
0.26 |
| chr4_106227685_106227836 | 0.23 |
ENSG00000243383 |
. |
12945 |
0.26 |
| chr2_43540601_43540961 | 0.23 |
AC010883.5 |
|
84069 |
0.09 |
| chr9_136923442_136923756 | 0.23 |
BRD3 |
bromodomain containing 3 |
2465 |
0.24 |
| chr7_14876053_14876204 | 0.23 |
DGKB |
diacylglycerol kinase, beta 90kDa |
4844 |
0.27 |
| chr17_71301605_71301911 | 0.23 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
5789 |
0.21 |
| chr4_26220381_26220779 | 0.23 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
53649 |
0.18 |
| chr13_36849620_36849771 | 0.23 |
ENSG00000207203 |
. |
8390 |
0.15 |
| chr17_68901911_68902062 | 0.23 |
ENSG00000238759 |
. |
392373 |
0.01 |
| chr7_101705458_101705808 | 0.22 |
CTA-357J21.1 |
|
29330 |
0.2 |
| chr7_4832075_4832390 | 0.22 |
ENSG00000264474 |
. |
3962 |
0.18 |
| chr2_48542892_48543532 | 0.22 |
FOXN2 |
forkhead box N2 |
1363 |
0.5 |
| chr3_187633047_187633241 | 0.22 |
BCL6 |
B-cell CLL/lymphoma 6 |
169629 |
0.03 |
| chr12_50362719_50362903 | 0.22 |
AQP6 |
aquaporin 6, kidney specific |
1789 |
0.21 |
| chr10_31073518_31073669 | 0.22 |
RP11-330O11.3 |
|
48723 |
0.15 |
| chr10_17256968_17257119 | 0.22 |
VIM-AS1 |
VIM antisense RNA 1 |
11854 |
0.15 |
| chr10_90984522_90984806 | 0.22 |
CH25H |
cholesterol 25-hydroxylase |
17593 |
0.15 |
| chr1_25198988_25199139 | 0.22 |
RUNX3 |
runt-related transcription factor 3 |
56549 |
0.12 |
| chr10_104474502_104474843 | 0.22 |
SFXN2 |
sideroflexin 2 |
279 |
0.68 |
| chr17_79481237_79481485 | 0.22 |
RP13-766D20.2 |
|
288 |
0.47 |
| chr7_23250461_23250612 | 0.22 |
NUPL2 |
nucleoporin like 2 |
28853 |
0.15 |
| chr22_46299735_46299886 | 0.22 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
68785 |
0.09 |
| chr4_48492543_48492694 | 0.22 |
ZAR1 |
zygote arrest 1 |
349 |
0.89 |
| chr5_14305370_14305521 | 0.22 |
TRIO |
trio Rho guanine nucleotide exchange factor |
14359 |
0.31 |
| chr2_27665478_27665738 | 0.22 |
KRTCAP3 |
keratinocyte associated protein 3 |
295 |
0.81 |
| chr12_18535849_18536000 | 0.22 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
100946 |
0.09 |
| chr11_67776489_67776640 | 0.22 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
516 |
0.66 |
| chr8_54117831_54118132 | 0.22 |
RP11-162D9.3 |
|
27834 |
0.21 |
| chr17_19234995_19235146 | 0.22 |
RP11-135L13.4 |
|
4370 |
0.13 |
| chr8_130472580_130472731 | 0.22 |
ENSG00000263763 |
. |
23733 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.2 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.2 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.1 | 0.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.2 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.1 | GO:1903504 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.2 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.2 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.2 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.5 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0032906 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.2 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0002604 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.0 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0010989 | regulation of low-density lipoprotein particle clearance(GO:0010988) negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 | 0.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.0 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0072501 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.0 | GO:0010984 | regulation of lipoprotein particle clearance(GO:0010984) |
| 0.0 | 0.0 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.5 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.0 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 0.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.0 | 0.0 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |