Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
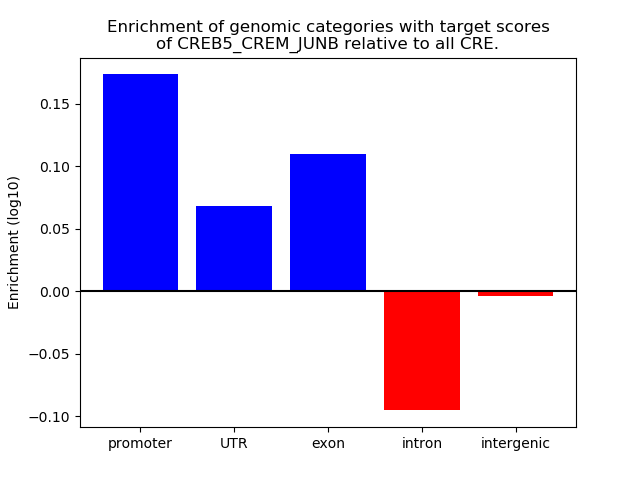
Results for CREB5_CREM_JUNB
Z-value: 0.95
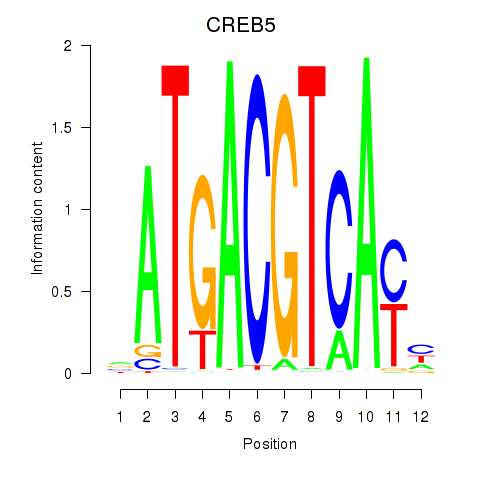
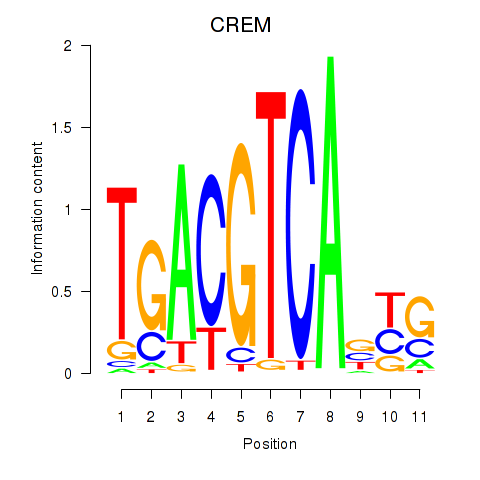
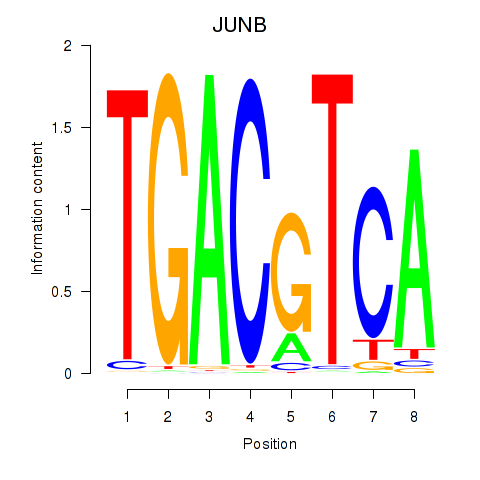
Transcription factors associated with CREB5_CREM_JUNB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB5
|
ENSG00000146592.12 | cAMP responsive element binding protein 5 |
|
CREM
|
ENSG00000095794.15 | cAMP responsive element modulator |
|
JUNB
|
ENSG00000171223.4 | JunB proto-oncogene, AP-1 transcription factor subunit |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_28725973_28726124 | CREB5 | 450 | 0.905693 | 0.85 | 4.0e-03 | Click! |
| chr7_28647428_28647579 | CREB5 | 1175 | 0.661254 | 0.80 | 9.3e-03 | Click! |
| chr7_28893632_28893840 | CREB5 | 44710 | 0.182883 | 0.79 | 1.1e-02 | Click! |
| chr7_28893326_28893622 | CREB5 | 44448 | 0.183745 | 0.78 | 1.3e-02 | Click! |
| chr7_28450467_28450674 | CREB5 | 1564 | 0.558990 | 0.77 | 1.5e-02 | Click! |
| chr10_35417017_35417356 | CREM | 781 | 0.469178 | 0.61 | 8.4e-02 | Click! |
| chr10_35418232_35418383 | CREM | 1902 | 0.239169 | -0.51 | 1.6e-01 | Click! |
| chr10_35454351_35454502 | CREM | 2018 | 0.269871 | -0.48 | 1.9e-01 | Click! |
| chr10_35438600_35438751 | CREM | 1375 | 0.361520 | 0.42 | 2.6e-01 | Click! |
| chr10_35484071_35484414 | CREM | 167 | 0.938050 | 0.38 | 3.2e-01 | Click! |
| chr19_12904796_12904947 | JUNB | 2561 | 0.094919 | 0.83 | 6.0e-03 | Click! |
| chr19_12901212_12902001 | JUNB | 704 | 0.380716 | 0.79 | 1.2e-02 | Click! |
| chr19_12903336_12903487 | JUNB | 1101 | 0.227887 | 0.71 | 3.4e-02 | Click! |
| chr19_12903631_12903782 | JUNB | 1396 | 0.172347 | 0.62 | 7.8e-02 | Click! |
| chr19_12902017_12902168 | JUNB | 218 | 0.805874 | 0.55 | 1.3e-01 | Click! |
Activity of the CREB5_CREM_JUNB motif across conditions
Conditions sorted by the z-value of the CREB5_CREM_JUNB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
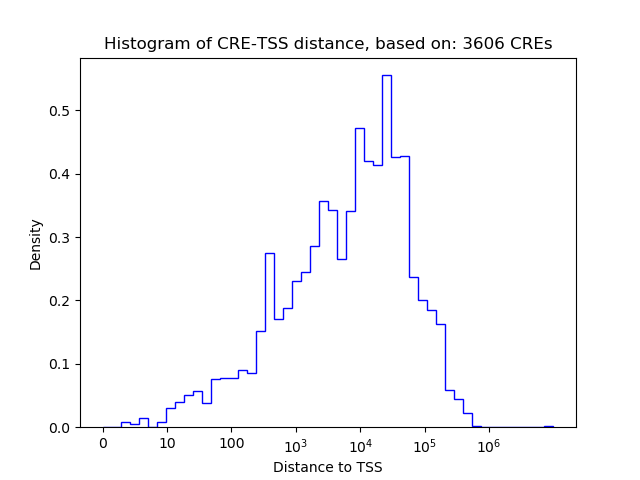
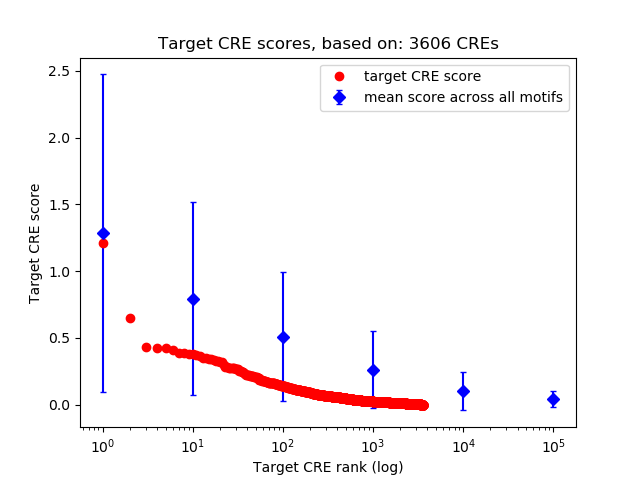
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_133022629_133022780 | 1.21 |
MUC8 |
mucin 8 |
28022 |
0.17 |
| chr1_11901364_11901515 | 0.65 |
NPPA-AS1 |
NPPA antisense RNA 1 |
365 |
0.78 |
| chr7_100782348_100782499 | 0.43 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
12044 |
0.1 |
| chr10_6959147_6959298 | 0.43 |
PRKCQ |
protein kinase C, theta |
336959 |
0.01 |
| chr4_153913148_153913299 | 0.42 |
FHDC1 |
FH2 domain containing 1 |
49088 |
0.16 |
| chr3_134092582_134092733 | 0.41 |
AMOTL2 |
angiomotin like 2 |
18 |
0.98 |
| chr11_12695755_12695947 | 0.39 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
118 |
0.98 |
| chr1_111033550_111033701 | 0.38 |
KCNA10 |
potassium voltage-gated channel, shaker-related subfamily, member 10 |
28172 |
0.13 |
| chr13_45343452_45343641 | 0.38 |
ENSG00000238932 |
. |
140737 |
0.04 |
| chr17_55446479_55446630 | 0.38 |
ENSG00000263902 |
. |
43008 |
0.17 |
| chr18_20840672_20840823 | 0.37 |
RP11-17J14.2 |
|
429 |
0.89 |
| chr15_94850174_94850340 | 0.37 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
8762 |
0.33 |
| chr8_49838453_49838744 | 0.35 |
SNAI2 |
snail family zinc finger 2 |
4299 |
0.35 |
| chr12_102872919_102873119 | 0.35 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
590 |
0.85 |
| chr18_22820121_22820315 | 0.35 |
ZNF521 |
zinc finger protein 521 |
8724 |
0.3 |
| chr6_80714655_80715350 | 0.34 |
TTK |
TTK protein kinase |
451 |
0.88 |
| chr2_38762873_38763024 | 0.34 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
34030 |
0.18 |
| chr1_161274764_161274974 | 0.33 |
MPZ |
myelin protein zero |
2392 |
0.18 |
| chr9_137348457_137348644 | 0.33 |
RXRA |
retinoid X receptor, alpha |
50122 |
0.15 |
| chr12_89511566_89511717 | 0.32 |
ENSG00000238302 |
. |
164421 |
0.04 |
| chr1_205216416_205216567 | 0.32 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
743 |
0.63 |
| chr19_35849271_35849508 | 0.29 |
FFAR3 |
free fatty acid receptor 3 |
27 |
0.95 |
| chr11_65256298_65256486 | 0.29 |
AP000769.1 |
Uncharacterized protein |
33664 |
0.08 |
| chr5_88566884_88567035 | 0.28 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
147665 |
0.05 |
| chrY_2576678_2576907 | 0.28 |
ENSG00000251841 |
. |
75998 |
0.11 |
| chr3_141514797_141515122 | 0.27 |
ENSG00000265391 |
. |
8206 |
0.2 |
| chr2_11888981_11889322 | 0.27 |
LPIN1 |
lipin 1 |
2387 |
0.21 |
| chr11_59279292_59279443 | 0.27 |
OR4D9 |
olfactory receptor, family 4, subfamily D, member 9 |
3019 |
0.16 |
| chr13_97876107_97876342 | 0.27 |
MBNL2 |
muscleblind-like splicing regulator 2 |
1615 |
0.53 |
| chr1_66999113_66999318 | 0.27 |
SGIP1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
60 |
0.98 |
| chr22_44569121_44569281 | 0.27 |
PARVG |
parvin, gamma |
365 |
0.92 |
| chr4_174198585_174198817 | 0.26 |
ENSG00000221296 |
. |
9390 |
0.17 |
| chr2_29537156_29537327 | 0.26 |
ALK |
anaplastic lymphoma receptor tyrosine kinase |
90981 |
0.08 |
| chr22_30662165_30662316 | 0.25 |
OSM |
oncostatin M |
433 |
0.7 |
| chr16_75587106_75587257 | 0.25 |
TMEM231 |
transmembrane protein 231 |
2955 |
0.16 |
| chr4_159131101_159131306 | 0.24 |
TMEM144 |
transmembrane protein 144 |
143 |
0.96 |
| chrX_2626678_2626926 | 0.24 |
CD99 |
CD99 molecule |
17261 |
0.2 |
| chr12_53784490_53784671 | 0.23 |
SP1 |
Sp1 transcription factor |
9494 |
0.12 |
| chr5_170739623_170739799 | 0.22 |
TLX3 |
T-cell leukemia homeobox 3 |
3423 |
0.23 |
| chr7_5492772_5492923 | 0.22 |
TNRC18 |
trinucleotide repeat containing 18 |
27802 |
0.1 |
| chr2_144280347_144280652 | 0.22 |
AC096558.1 |
|
42141 |
0.16 |
| chr12_19389657_19389808 | 0.22 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
126 |
0.97 |
| chr1_2216519_2216670 | 0.22 |
RP4-713A8.1 |
|
41538 |
0.09 |
| chr7_138393128_138393279 | 0.22 |
SVOPL |
SVOP-like |
7106 |
0.24 |
| chr6_14477462_14477818 | 0.22 |
ENSG00000206960 |
. |
169126 |
0.04 |
| chr9_123346254_123346525 | 0.21 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
3952 |
0.35 |
| chr6_53213778_53213929 | 0.21 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
48 |
0.98 |
| chr9_130912677_130912874 | 0.21 |
LCN2 |
lipocalin 2 |
978 |
0.34 |
| chr15_98224143_98224294 | 0.21 |
LINC00923 |
long intergenic non-protein coding RNA 923 |
193562 |
0.03 |
| chr15_49336349_49337349 | 0.21 |
SECISBP2L |
SECIS binding protein 2-like |
1781 |
0.28 |
| chr4_113298543_113298694 | 0.20 |
ALPK1 |
alpha-kinase 1 |
80082 |
0.08 |
| chr2_28114153_28114304 | 0.20 |
RBKS |
ribokinase |
263 |
0.74 |
| chr2_95745044_95745195 | 0.20 |
AC103563.9 |
|
26198 |
0.14 |
| chr4_186316955_186317190 | 0.20 |
LRP2BP |
LRP2 binding protein |
19 |
0.61 |
| chr17_75531537_75531688 | 0.19 |
SEPT9 |
septin 9 |
53617 |
0.12 |
| chr6_119417895_119418107 | 0.19 |
FAM184A |
family with sequence similarity 184, member A |
17657 |
0.19 |
| chr21_18983750_18984027 | 0.18 |
BTG3 |
BTG family, member 3 |
1274 |
0.5 |
| chr18_46557895_46558135 | 0.18 |
RP11-15F12.1 |
|
7942 |
0.2 |
| chr3_50337208_50337424 | 0.18 |
HYAL3 |
hyaluronoglucosaminidase 3 |
417 |
0.42 |
| chr1_231173648_231173799 | 0.18 |
FAM89A |
family with sequence similarity 89, member A |
2269 |
0.26 |
| chr17_27889238_27889394 | 0.18 |
RP11-68I3.2 |
|
1751 |
0.18 |
| chr2_8552253_8552511 | 0.18 |
AC011747.7 |
|
263514 |
0.02 |
| chr1_22380057_22380208 | 0.18 |
CDC42 |
cell division cycle 42 |
342 |
0.82 |
| chr12_62861008_62861159 | 0.17 |
MON2 |
MON2 homolog (S. cerevisiae) |
454 |
0.83 |
| chr1_236229504_236229679 | 0.17 |
NID1 |
nidogen 1 |
1129 |
0.52 |
| chr2_160471369_160471526 | 0.17 |
AC009506.1 |
|
360 |
0.68 |
| chr16_57770832_57771049 | 0.17 |
KATNB1 |
katanin p80 (WD repeat containing) subunit B 1 |
55 |
0.96 |
| chr16_48428377_48428528 | 0.17 |
SIAH1 |
siah E3 ubiquitin protein ligase 1 |
586 |
0.72 |
| chr19_996464_996712 | 0.17 |
AC004528.1 |
Uncharacterized protein |
3013 |
0.1 |
| chrY_6954060_6954211 | 0.17 |
TBL1Y |
transducin (beta)-like 1, Y-linked |
175408 |
0.03 |
| chr5_73730230_73730381 | 0.16 |
ENSG00000244326 |
. |
116494 |
0.06 |
| chr12_54446931_54447133 | 0.16 |
HOXC4 |
homeobox C4 |
629 |
0.5 |
| chr12_108958034_108958185 | 0.16 |
ISCU |
iron-sulfur cluster assembly enzyme |
1723 |
0.27 |
| chr6_158508122_158508351 | 0.16 |
SYNJ2 |
synaptojanin 2 |
9524 |
0.2 |
| chr8_65490045_65490217 | 0.16 |
RP11-21C4.1 |
|
311 |
0.91 |
| chr4_8430147_8430298 | 0.16 |
ACOX3 |
acyl-CoA oxidase 3, pristanoyl |
9 |
0.98 |
| chr18_10791557_10791762 | 0.16 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
11 |
0.99 |
| chr10_43474933_43475084 | 0.16 |
ENSG00000263795 |
. |
18003 |
0.25 |
| chr9_86323490_86324000 | 0.16 |
RP11-522I20.3 |
|
512 |
0.52 |
| chr16_16047180_16047331 | 0.16 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
3821 |
0.26 |
| chr17_42423914_42424082 | 0.16 |
GRN |
granulin |
1325 |
0.25 |
| chr19_38908795_38908946 | 0.16 |
RASGRP4 |
RAS guanyl releasing protein 4 |
7932 |
0.09 |
| chr8_6114669_6114820 | 0.16 |
RP11-115C21.2 |
|
149319 |
0.04 |
| chr14_101292334_101292485 | 0.15 |
AL117190.2 |
|
3128 |
0.07 |
| chr18_74205447_74205647 | 0.15 |
ZNF516 |
zinc finger protein 516 |
1599 |
0.27 |
| chr21_44937001_44937451 | 0.15 |
SIK1 |
salt-inducible kinase 1 |
90218 |
0.08 |
| chr15_40636361_40636512 | 0.15 |
C15orf52 |
chromosome 15 open reading frame 52 |
3268 |
0.1 |
| chr2_218027426_218027577 | 0.15 |
ENSG00000251849 |
. |
13720 |
0.27 |
| chr14_71246503_71246654 | 0.15 |
MAP3K9 |
mitogen-activated protein kinase kinase kinase 9 |
3579 |
0.34 |
| chr7_1979584_1979735 | 0.15 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
526 |
0.85 |
| chr2_102093131_102093315 | 0.15 |
RFX8 |
RFX family member 8, lacking RFX DNA binding domain |
2058 |
0.43 |
| chr6_90929386_90929701 | 0.15 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
76918 |
0.1 |
| chr9_134516940_134517091 | 0.15 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
15680 |
0.22 |
| chr2_11838198_11838349 | 0.15 |
AC106875.1 |
|
16686 |
0.12 |
| chr1_32810829_32811029 | 0.14 |
TSSK3 |
testis-specific serine kinase 3 |
8126 |
0.12 |
| chr20_33146664_33146909 | 0.14 |
MAP1LC3A |
microtubule-associated protein 1 light chain 3 alpha |
264 |
0.91 |
| chr6_138126801_138126952 | 0.14 |
RP11-356I2.4 |
|
20155 |
0.19 |
| chr21_36206001_36206152 | 0.14 |
RUNX1 |
runt-related transcription factor 1 |
53404 |
0.16 |
| chr20_45934513_45934664 | 0.14 |
ENSG00000265532 |
. |
2538 |
0.2 |
| chr17_65505786_65506259 | 0.14 |
CTD-2653B5.1 |
|
14575 |
0.17 |
| chr22_39163031_39163182 | 0.14 |
SUN2 |
Sad1 and UNC84 domain containing 2 |
11101 |
0.08 |
| chr6_2449367_2449525 | 0.14 |
ENSG00000266252 |
. |
39577 |
0.21 |
| chr5_70751007_70751459 | 0.14 |
BDP1 |
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
209 |
0.96 |
| chr1_17630639_17630914 | 0.14 |
PADI4 |
peptidyl arginine deiminase, type IV |
3914 |
0.2 |
| chr17_72783237_72783388 | 0.14 |
TMEM104 |
transmembrane protein 104 |
10653 |
0.11 |
| chr19_8454744_8454908 | 0.14 |
RAB11B |
RAB11B, member RAS oncogene family |
39 |
0.64 |
| chr1_28099274_28099549 | 0.13 |
STX12 |
syntaxin 12 |
283 |
0.78 |
| chr16_73072287_73072438 | 0.13 |
ZFHX3 |
zinc finger homeobox 3 |
9912 |
0.21 |
| chr5_66492437_66492594 | 0.13 |
CD180 |
CD180 molecule |
112 |
0.98 |
| chr3_128195185_128195336 | 0.13 |
DNAJB8 |
DnaJ (Hsp40) homolog, subfamily B, member 8 |
9169 |
0.15 |
| chr16_22825426_22825577 | 0.13 |
HS3ST2 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
3 |
0.99 |
| chr2_65309092_65309434 | 0.13 |
RAB1A |
RAB1A, member RAS oncogene family |
24261 |
0.15 |
| chr2_70750633_70750894 | 0.13 |
TGFA |
transforming growth factor, alpha |
29859 |
0.16 |
| chr5_64331605_64331756 | 0.13 |
ENSG00000207439 |
. |
87516 |
0.1 |
| chr20_62580456_62580607 | 0.13 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
1948 |
0.13 |
| chr8_125726263_125726555 | 0.13 |
MTSS1 |
metastasis suppressor 1 |
13448 |
0.25 |
| chr4_141015090_141015375 | 0.13 |
RP11-392B6.1 |
|
33937 |
0.2 |
| chr7_132095239_132095390 | 0.13 |
AC011625.1 |
|
58221 |
0.15 |
| chr21_36295704_36295877 | 0.13 |
RUNX1 |
runt-related transcription factor 1 |
33703 |
0.24 |
| chr1_206235900_206236051 | 0.13 |
AVPR1B |
arginine vasopressin receptor 1B |
11999 |
0.16 |
| chr1_15433135_15433286 | 0.12 |
TMEM51 |
transmembrane protein 51 |
45818 |
0.16 |
| chr2_171210862_171211026 | 0.12 |
AC012594.1 |
|
3316 |
0.37 |
| chr4_154713480_154713740 | 0.12 |
SFRP2 |
secreted frizzled-related protein 2 |
3338 |
0.26 |
| chr15_70821207_70821700 | 0.12 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
173167 |
0.03 |
| chr16_58535345_58535501 | 0.12 |
NDRG4 |
NDRG family member 4 |
16 |
0.97 |
| chr19_4584744_4584943 | 0.12 |
SEMA6B |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
25023 |
0.09 |
| chr9_114288073_114288445 | 0.12 |
ZNF483 |
zinc finger protein 483 |
746 |
0.65 |
| chr12_78813400_78813551 | 0.12 |
NAV3 |
neuron navigator 3 |
220619 |
0.02 |
| chr8_146012726_146012877 | 0.12 |
ZNF34 |
zinc finger protein 34 |
71 |
0.94 |
| chr21_43673739_43673890 | 0.12 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
2380 |
0.26 |
| chr19_44087068_44087219 | 0.12 |
PINLYP |
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
1954 |
0.14 |
| chr10_43429375_43429752 | 0.12 |
ENSG00000263795 |
. |
63448 |
0.12 |
| chr7_142216694_142216845 | 0.12 |
MTRNR2L6 |
MT-RNR2-like 6 |
157335 |
0.04 |
| chr6_108486750_108487194 | 0.12 |
OSTM1 |
osteopetrosis associated transmembrane protein 1 |
72 |
0.78 |
| chr20_30192135_30192286 | 0.12 |
ID1 |
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
876 |
0.44 |
| chr9_78990142_78990293 | 0.12 |
RFK |
riboflavin kinase |
18831 |
0.24 |
| chr14_75801482_75801633 | 0.11 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
54661 |
0.1 |
| chr15_80365699_80365932 | 0.11 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
883 |
0.66 |
| chr10_121580538_121580689 | 0.11 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
2385 |
0.32 |
| chr1_151254019_151254394 | 0.11 |
ZNF687 |
zinc finger protein 687 |
112 |
0.61 |
| chr2_44345424_44345575 | 0.11 |
ENSG00000252599 |
. |
36535 |
0.15 |
| chr13_113664073_113664237 | 0.11 |
RP11-120K24.5 |
|
170 |
0.9 |
| chr19_6552264_6552452 | 0.11 |
ENSG00000199223 |
. |
20710 |
0.1 |
| chr2_110013073_110013224 | 0.11 |
ENSG00000265965 |
. |
83067 |
0.11 |
| chrX_129318276_129318427 | 0.11 |
RAB33A |
RAB33A, member RAS oncogene family |
12728 |
0.19 |
| chr2_66723271_66723422 | 0.11 |
MEIS1 |
Meis homeobox 1 |
12713 |
0.22 |
| chr3_62304486_62304637 | 0.11 |
ENSG00000241472 |
. |
6 |
0.57 |
| chr2_111453217_111453510 | 0.11 |
BUB1 |
BUB1 mitotic checkpoint serine/threonine kinase |
17672 |
0.21 |
| chr2_1609070_1609221 | 0.11 |
AC144450.1 |
|
14740 |
0.26 |
| chr18_60859816_60859967 | 0.11 |
ENSG00000238988 |
. |
2007 |
0.39 |
| chr2_197944933_197945147 | 0.11 |
ANKRD44 |
ankyrin repeat domain 44 |
30528 |
0.22 |
| chr10_71992203_71992435 | 0.11 |
PPA1 |
pyrophosphatase (inorganic) 1 |
841 |
0.65 |
| chr10_12379442_12379593 | 0.11 |
ENSG00000264036 |
. |
9129 |
0.18 |
| chr9_137263810_137263961 | 0.11 |
ENSG00000263897 |
. |
7372 |
0.25 |
| chr6_44355096_44355314 | 0.11 |
CDC5L |
cell division cycle 5-like |
57 |
0.97 |
| chr2_219434859_219435206 | 0.11 |
RQCD1 |
RCD1 required for cell differentiation1 homolog (S. pombe) |
1434 |
0.25 |
| chr1_151809393_151809571 | 0.11 |
C2CD4D |
C2 calcium-dependent domain containing 4D |
3551 |
0.1 |
| chr2_62520408_62520559 | 0.11 |
ENSG00000238809 |
. |
28353 |
0.17 |
| chr1_167193026_167193202 | 0.11 |
POU2F1 |
POU class 2 homeobox 1 |
2971 |
0.23 |
| chrX_83757427_83757578 | 0.11 |
HDX |
highly divergent homeobox |
15 |
0.51 |
| chr4_174443398_174443549 | 0.10 |
HAND2-AS1 |
HAND2 antisense RNA 1 (head to head) |
4948 |
0.23 |
| chr14_102785720_102785871 | 0.10 |
ZNF839 |
zinc finger protein 839 |
160 |
0.94 |
| chr20_17207212_17207363 | 0.10 |
PCSK2 |
proprotein convertase subtilisin/kexin type 2 |
349 |
0.9 |
| chr2_47213208_47213406 | 0.10 |
RP11-15I20.1 |
|
29280 |
0.14 |
| chr1_172189218_172189369 | 0.10 |
DNM3OS |
DNM3 opposite strand/antisense RNA |
75359 |
0.09 |
| chr3_122513525_122513727 | 0.10 |
DIRC2 |
disrupted in renal carcinoma 2 |
16 |
0.96 |
| chr2_203735601_203735829 | 0.10 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
54 |
0.98 |
| chr3_8543150_8543354 | 0.10 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
32 |
0.64 |
| chr19_49142340_49142538 | 0.10 |
CA11 |
carbonic anhydrase XI |
277 |
0.77 |
| chr2_8144515_8144736 | 0.10 |
ENSG00000221255 |
. |
427653 |
0.01 |
| chr2_28837076_28837338 | 0.10 |
PLB1 |
phospholipase B1 |
12421 |
0.21 |
| chr3_185080983_185081411 | 0.10 |
MAP3K13 |
mitogen-activated protein kinase kinase kinase 13 |
289 |
0.93 |
| chr18_3712580_3712731 | 0.10 |
RP11-874J12.3 |
|
58775 |
0.1 |
| chr8_67343364_67343686 | 0.10 |
ADHFE1 |
alcohol dehydrogenase, iron containing, 1 |
1105 |
0.48 |
| chr3_115231996_115232281 | 0.10 |
GAP43 |
growth associated protein 43 |
110033 |
0.07 |
| chr8_38385040_38385321 | 0.10 |
C8orf86 |
chromosome 8 open reading frame 86 |
995 |
0.59 |
| chr14_81736950_81737101 | 0.10 |
STON2 |
stonin 2 |
6252 |
0.26 |
| chr10_1296680_1296831 | 0.10 |
ADARB2 |
adenosine deaminase, RNA-specific, B2 (non-functional) |
50430 |
0.16 |
| chr7_138914973_138915166 | 0.10 |
UBN2 |
ubinuclein 2 |
33 |
0.98 |
| chr12_4229354_4229505 | 0.10 |
CCND2 |
cyclin D2 |
153509 |
0.04 |
| chr5_138738832_138739361 | 0.10 |
SPATA24 |
spermatogenesis associated 24 |
643 |
0.54 |
| chr7_101723621_101723772 | 0.10 |
CTA-357J21.1 |
|
47393 |
0.15 |
| chr2_15282512_15282709 | 0.10 |
NBAS |
neuroblastoma amplified sequence |
47911 |
0.2 |
| chr2_196531011_196531162 | 0.10 |
ENSG00000201813 |
. |
4112 |
0.27 |
| chr3_32451109_32451284 | 0.10 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
17665 |
0.23 |
| chr6_14094353_14094642 | 0.10 |
CD83 |
CD83 molecule |
23375 |
0.24 |
| chr11_14994041_14994192 | 0.10 |
CALCA |
calcitonin-related polypeptide alpha |
216 |
0.96 |
| chr3_142995226_142995377 | 0.10 |
RP13-635I23.3 |
|
37970 |
0.17 |
| chr7_6183718_6183869 | 0.10 |
USP42 |
ubiquitin specific peptidase 42 |
27682 |
0.11 |
| chr10_22629149_22629300 | 0.10 |
SPAG6 |
sperm associated antigen 6 |
5175 |
0.17 |
| chr7_26206357_26206508 | 0.09 |
CTB-119C2.1 |
|
8133 |
0.18 |
| chr5_179453537_179453707 | 0.09 |
ENSG00000198995 |
. |
11225 |
0.19 |
| chr7_95916382_95916744 | 0.09 |
ENSG00000220987 |
. |
19609 |
0.16 |
| chr2_64957617_64958033 | 0.09 |
ENSG00000253082 |
. |
45413 |
0.14 |
| chr12_56731704_56731855 | 0.09 |
IL23A |
interleukin 23, alpha subunit p19 |
884 |
0.33 |
| chr1_243607561_243607712 | 0.09 |
ENSG00000265201 |
. |
98158 |
0.07 |
| chr16_87469163_87469969 | 0.09 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
2826 |
0.22 |
| chr15_84977138_84977289 | 0.09 |
UBE2Q2P11 |
ubiquitin-conjugating enzyme E2Q family member 2 pseudogene 11 |
678 |
0.57 |
| chr11_930105_930418 | 0.09 |
AP2A2 |
adaptor-related protein complex 2, alpha 2 subunit |
2432 |
0.17 |
| chr17_18528921_18529072 | 0.09 |
CCDC144B |
coiled-coil domain containing 144B (pseudogene) |
75 |
0.96 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.1 | 0.2 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.1 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.3 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0031272 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.0 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.3 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.0 | GO:0032634 | interleukin-5 production(GO:0032634) regulation of interleukin-5 production(GO:0032674) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.3 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |