Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
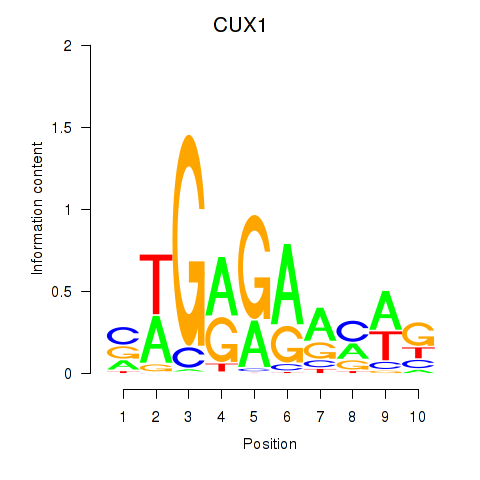
Results for CUX1
Z-value: 0.72
Transcription factors associated with CUX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX1
|
ENSG00000257923.5 | cut like homeobox 1 |
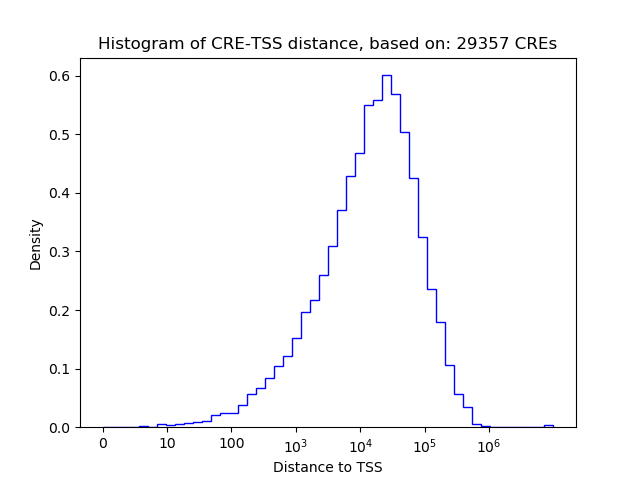
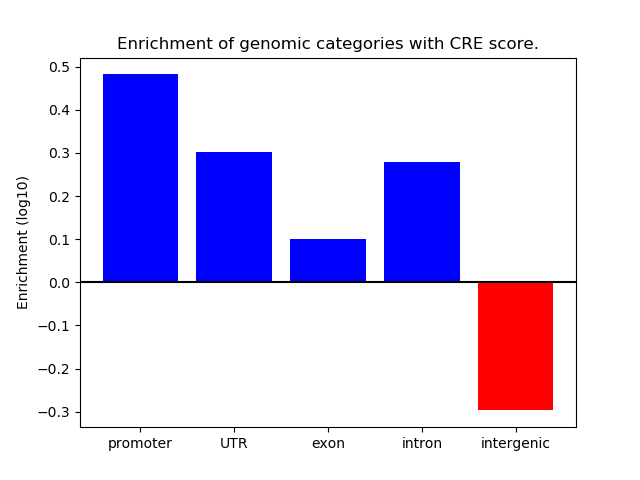
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_101388868_101389019 | CUX1 | 70016 | 0.105292 | 0.73 | 2.6e-02 | Click! |
| chr7_101378437_101378588 | CUX1 | 80447 | 0.089167 | 0.66 | 5.5e-02 | Click! |
| chr7_101438358_101438509 | CUX1 | 20526 | 0.207993 | -0.60 | 8.5e-02 | Click! |
| chr7_101376791_101377001 | CUX1 | 82063 | 0.086907 | 0.57 | 1.1e-01 | Click! |
| chr7_101387918_101388069 | CUX1 | 70966 | 0.103709 | 0.56 | 1.2e-01 | Click! |
Activity of the CUX1 motif across conditions
Conditions sorted by the z-value of the CUX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
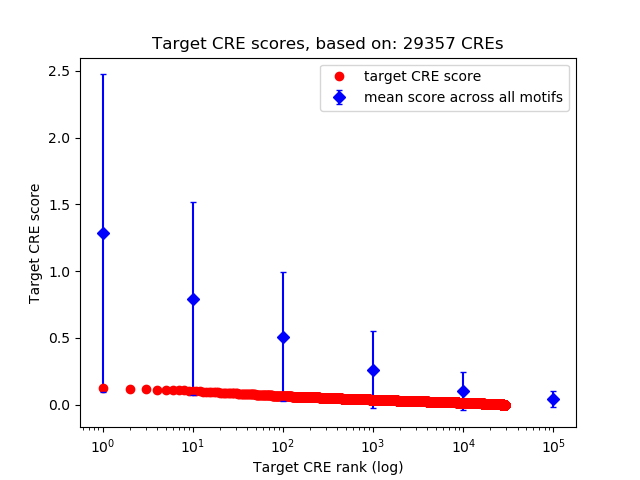
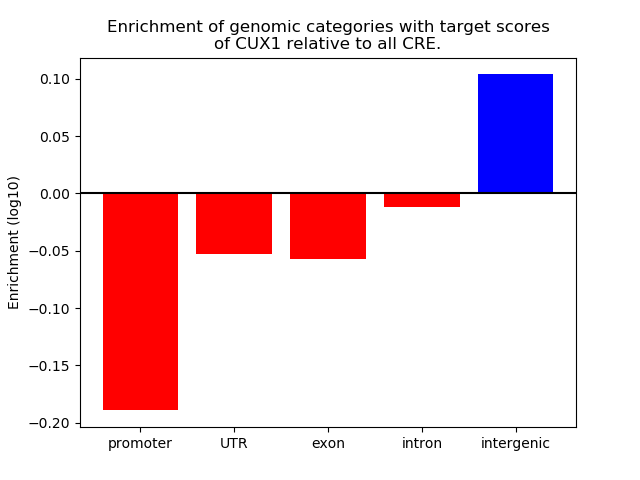
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr20_4356829_4357115 | 0.12 |
ADRA1D |
adrenoceptor alpha 1D |
127251 |
0.05 |
| chr6_89504471_89504714 | 0.12 |
ENSG00000222145 |
. |
80443 |
0.1 |
| chr5_102780141_102780414 | 0.12 |
NUDT12 |
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
118213 |
0.06 |
| chr5_77887462_77888069 | 0.11 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
42791 |
0.21 |
| chr7_114628946_114629097 | 0.11 |
MDFIC |
MyoD family inhibitor domain containing |
55097 |
0.17 |
| chr22_20777294_20777549 | 0.11 |
ENSG00000207343 |
. |
6232 |
0.12 |
| chr10_33529675_33530002 | 0.11 |
NRP1 |
neuropilin 1 |
22794 |
0.19 |
| chr7_100411285_100411882 | 0.11 |
ENSG00000263672 |
. |
7303 |
0.11 |
| chr14_77511448_77512058 | 0.11 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
16719 |
0.18 |
| chr12_66322607_66323042 | 0.10 |
AC090673.2 |
Uncharacterized protein |
4857 |
0.21 |
| chr15_39696001_39696152 | 0.10 |
RP11-624L4.1 |
|
16834 |
0.27 |
| chr2_113594705_113594960 | 0.10 |
IL1B |
interleukin 1, beta |
352 |
0.86 |
| chr21_40378291_40378490 | 0.10 |
ENSG00000272015 |
. |
111681 |
0.06 |
| chr21_47485945_47486454 | 0.10 |
AP001471.1 |
|
31245 |
0.13 |
| chr4_186651858_186652115 | 0.10 |
SORBS2 |
sorbin and SH3 domain containing 2 |
9498 |
0.21 |
| chr5_159107_159571 | 0.09 |
ENSG00000199540 |
. |
15027 |
0.14 |
| chr7_54878481_54878866 | 0.09 |
SEC61G |
Sec61 gamma subunit |
51006 |
0.13 |
| chr11_73174890_73175041 | 0.09 |
RP11-809N8.4 |
|
58623 |
0.1 |
| chr6_136434664_136434859 | 0.09 |
PDE7B |
phosphodiesterase 7B |
35463 |
0.16 |
| chr9_110037719_110038006 | 0.09 |
RAD23B |
RAD23 homolog B (S. cerevisiae) |
7556 |
0.31 |
| chr1_245951430_245951581 | 0.09 |
RP11-522M21.3 |
|
111725 |
0.07 |
| chr1_110476345_110476632 | 0.09 |
CSF1 |
colony stimulating factor 1 (macrophage) |
22880 |
0.15 |
| chr6_130644268_130644419 | 0.09 |
SAMD3 |
sterile alpha motif domain containing 3 |
42227 |
0.17 |
| chr11_128442939_128443342 | 0.09 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
14313 |
0.23 |
| chr9_16031542_16031875 | 0.09 |
C9orf92 |
chromosome 9 open reading frame 92 |
184189 |
0.03 |
| chr20_11229977_11230245 | 0.09 |
C20orf187 |
chromosome 20 open reading frame 187 |
221300 |
0.02 |
| chr17_26793910_26794378 | 0.09 |
SLC13A2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
6167 |
0.11 |
| chr12_6198255_6198797 | 0.09 |
ENSG00000240533 |
. |
6610 |
0.23 |
| chr18_46354811_46355214 | 0.09 |
RP11-484L8.1 |
|
6129 |
0.29 |
| chr7_2867050_2867285 | 0.08 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
12274 |
0.25 |
| chr7_132004161_132004312 | 0.08 |
AC011625.1 |
|
32857 |
0.21 |
| chr2_48795884_48796035 | 0.08 |
STON1-GTF2A1L |
STON1-GTF2A1L readthrough |
200 |
0.95 |
| chr10_45476507_45476680 | 0.08 |
C10orf10 |
chromosome 10 open reading frame 10 |
2335 |
0.19 |
| chr12_95597054_95597379 | 0.08 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
13939 |
0.18 |
| chr2_121005547_121005698 | 0.08 |
RALB |
v-ral simian leukemia viral oncogene homolog B |
4712 |
0.21 |
| chr4_140933484_140933746 | 0.08 |
RP11-392B6.1 |
|
115554 |
0.06 |
| chr18_10479454_10479605 | 0.08 |
APCDD1 |
adenomatosis polyposis coli down-regulated 1 |
7696 |
0.26 |
| chr1_66478233_66478531 | 0.08 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
20277 |
0.22 |
| chr10_103295849_103296000 | 0.08 |
DPCD |
deleted in primary ciliary dyskinesia homolog (mouse) |
34482 |
0.14 |
| chr10_88162815_88162966 | 0.08 |
GRID1 |
glutamate receptor, ionotropic, delta 1 |
36655 |
0.16 |
| chr3_38871334_38871622 | 0.08 |
RP11-134J21.1 |
|
6085 |
0.2 |
| chr10_112182641_112182792 | 0.08 |
ENSG00000221359 |
. |
11283 |
0.22 |
| chr5_94742657_94743120 | 0.08 |
FAM81B |
family with sequence similarity 81, member B |
15840 |
0.25 |
| chr9_131905468_131905894 | 0.08 |
PPP2R4 |
protein phosphatase 2A activator, regulatory subunit 4 |
1377 |
0.3 |
| chr8_71060509_71060843 | 0.08 |
NCOA2 |
nuclear receptor coactivator 2 |
19 |
0.98 |
| chr19_54023625_54024053 | 0.08 |
ZNF331 |
zinc finger protein 331 |
439 |
0.74 |
| chr2_30834318_30834573 | 0.08 |
LCLAT1 |
lysocardiolipin acyltransferase 1 |
164176 |
0.04 |
| chr2_216765021_216765281 | 0.08 |
ENSG00000212055 |
. |
21509 |
0.25 |
| chr22_22191485_22191760 | 0.08 |
MAPK1 |
mitogen-activated protein kinase 1 |
30108 |
0.12 |
| chr9_16540961_16541659 | 0.08 |
RP11-183I6.2 |
|
68124 |
0.12 |
| chr6_25168636_25168787 | 0.08 |
ENSG00000222373 |
. |
23930 |
0.17 |
| chr17_56582578_56582941 | 0.08 |
MTMR4 |
myotubularin related protein 4 |
8638 |
0.12 |
| chr7_148410320_148410484 | 0.08 |
CUL1 |
cullin 1 |
14398 |
0.17 |
| chr10_114719872_114720172 | 0.08 |
RP11-57H14.2 |
|
8388 |
0.22 |
| chr9_117805131_117805394 | 0.08 |
TNC |
tenascin C |
21929 |
0.25 |
| chr10_75924551_75924986 | 0.08 |
ADK |
adenosine kinase |
11676 |
0.2 |
| chr18_43381728_43382214 | 0.07 |
SIGLEC15 |
sialic acid binding Ig-like lectin 15 |
23506 |
0.18 |
| chr1_203020776_203021050 | 0.07 |
PPFIA4 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
545 |
0.7 |
| chr16_1521079_1521553 | 0.07 |
CLCN7 |
chloride channel, voltage-sensitive 7 |
641 |
0.43 |
| chr2_177508527_177508764 | 0.07 |
ENSG00000252027 |
. |
20759 |
0.27 |
| chr5_94423327_94423550 | 0.07 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
5850 |
0.32 |
| chr3_10018102_10018700 | 0.07 |
EMC3 |
ER membrane protein complex subunit 3 |
9965 |
0.08 |
| chr3_177549479_177549630 | 0.07 |
ENSG00000199858 |
. |
176013 |
0.03 |
| chr10_97047482_97047633 | 0.07 |
PDLIM1 |
PDZ and LIM domain 1 |
3224 |
0.31 |
| chr20_44470840_44471081 | 0.07 |
SNX21 |
sorting nexin family member 21 |
8185 |
0.08 |
| chr6_109663929_109664175 | 0.07 |
ENSG00000201023 |
. |
37355 |
0.11 |
| chr14_91305961_91306112 | 0.07 |
TTC7B |
tetratricopeptide repeat domain 7B |
23213 |
0.24 |
| chr15_59588622_59588914 | 0.07 |
RP11-429D19.1 |
|
25407 |
0.13 |
| chr2_8722437_8722588 | 0.07 |
AC011747.7 |
|
93384 |
0.09 |
| chr9_94154220_94154371 | 0.07 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
30100 |
0.21 |
| chr7_101360762_101360997 | 0.07 |
MYL10 |
myosin, light chain 10, regulatory |
88303 |
0.08 |
| chr4_74844819_74845072 | 0.07 |
PF4 |
platelet factor 4 |
2896 |
0.19 |
| chr6_7151631_7152228 | 0.07 |
RREB1 |
ras responsive element binding protein 1 |
12451 |
0.19 |
| chr12_63253482_63253831 | 0.07 |
ENSG00000200296 |
. |
8975 |
0.26 |
| chr4_72271640_72271791 | 0.07 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
66945 |
0.14 |
| chr7_26714510_26714661 | 0.07 |
C7orf71 |
chromosome 7 open reading frame 71 |
37095 |
0.2 |
| chr13_27543257_27543411 | 0.07 |
USP12-AS1 |
USP12 antisense RNA 1 |
193658 |
0.02 |
| chr10_14654331_14654637 | 0.07 |
FAM107B |
family with sequence similarity 107, member B |
67 |
0.98 |
| chr1_235243157_235243394 | 0.07 |
ENSG00000207181 |
. |
47977 |
0.13 |
| chr9_110905251_110905592 | 0.07 |
ENSG00000222512 |
. |
215788 |
0.02 |
| chr7_47541782_47542068 | 0.07 |
TNS3 |
tensin 3 |
21042 |
0.28 |
| chr1_81698308_81698583 | 0.07 |
ENSG00000223026 |
. |
19029 |
0.27 |
| chr10_3793162_3793446 | 0.07 |
RP11-184A2.3 |
|
45 |
0.98 |
| chr2_62638288_62638545 | 0.07 |
ENSG00000241625 |
. |
79897 |
0.09 |
| chr2_171219129_171219280 | 0.07 |
AC012594.1 |
|
11576 |
0.29 |
| chr7_149666181_149666332 | 0.07 |
ATP6V0E2-AS1 |
ATP6V0E2 antisense RNA 1 |
88557 |
0.08 |
| chr1_9950424_9950885 | 0.07 |
CTNNBIP1 |
catenin, beta interacting protein 1 |
2641 |
0.21 |
| chr2_224599920_224600071 | 0.07 |
AP1S3 |
adaptor-related protein complex 1, sigma 3 subunit |
102206 |
0.08 |
| chr2_192241823_192241993 | 0.07 |
ENSG00000252130 |
. |
6244 |
0.26 |
| chr8_101450194_101450600 | 0.07 |
KB-1615E4.2 |
|
37506 |
0.14 |
| chr2_43188272_43188475 | 0.07 |
ENSG00000207087 |
. |
130259 |
0.05 |
| chr11_85658543_85658752 | 0.07 |
RP11-90K17.2 |
|
23922 |
0.18 |
| chr4_140669620_140669850 | 0.07 |
ENSG00000252233 |
. |
45852 |
0.14 |
| chr6_42105086_42105410 | 0.07 |
C6orf132 |
chromosome 6 open reading frame 132 |
4934 |
0.17 |
| chr1_85245912_85246495 | 0.07 |
ENSG00000251899 |
. |
12679 |
0.24 |
| chr5_39411390_39411705 | 0.07 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
13423 |
0.26 |
| chr1_154703230_154703438 | 0.07 |
ADAR |
adenosine deaminase, RNA-specific |
102860 |
0.05 |
| chr10_128111271_128111422 | 0.07 |
ADAM12 |
ADAM metallopeptidase domain 12 |
34322 |
0.19 |
| chr3_128690038_128690295 | 0.07 |
RP11-723O4.6 |
Uncharacterized protein FLJ43738 |
7 |
0.97 |
| chr10_72546347_72546498 | 0.07 |
TBATA |
thymus, brain and testes associated |
1265 |
0.51 |
| chr3_138493910_138494448 | 0.07 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
4782 |
0.31 |
| chr12_13192614_13192765 | 0.07 |
KIAA1467 |
KIAA1467 |
4529 |
0.19 |
| chr9_96902563_96902834 | 0.07 |
ENSG00000199165 |
. |
35536 |
0.13 |
| chr2_207387205_207387356 | 0.06 |
ADAM23 |
ADAM metallopeptidase domain 23 |
72279 |
0.1 |
| chr3_141507253_141507404 | 0.06 |
ENSG00000265391 |
. |
575 |
0.76 |
| chr17_56454718_56454869 | 0.06 |
SUPT4H1 |
suppressor of Ty 4 homolog 1 (S. cerevisiae) |
24339 |
0.1 |
| chr2_106088158_106088309 | 0.06 |
FHL2 |
four and a half LIM domains 2 |
33263 |
0.2 |
| chr18_10059619_10060204 | 0.06 |
ENSG00000263630 |
. |
54717 |
0.15 |
| chr9_7368414_7368565 | 0.06 |
KDM4C |
lysine (K)-specific demethylase 4C |
354701 |
0.01 |
| chr2_47284996_47285404 | 0.06 |
AC073283.7 |
|
9761 |
0.19 |
| chr22_32272417_32272584 | 0.06 |
DEPDC5 |
DEP domain containing 5 |
23598 |
0.15 |
| chr10_43624518_43624697 | 0.06 |
CSGALNACT2 |
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
9327 |
0.24 |
| chr3_72536153_72536343 | 0.06 |
RYBP |
RING1 and YY1 binding protein |
40179 |
0.18 |
| chr1_90289087_90289450 | 0.06 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
1788 |
0.4 |
| chr1_161691025_161691176 | 0.06 |
FCRLB |
Fc receptor-like B |
253 |
0.86 |
| chr4_157466044_157466305 | 0.06 |
RP11-171N4.2 |
Uncharacterized protein |
97285 |
0.08 |
| chr16_57798443_57798674 | 0.06 |
KIFC3 |
kinesin family member C3 |
257 |
0.89 |
| chr12_11733293_11733560 | 0.06 |
ENSG00000251747 |
. |
34013 |
0.16 |
| chr7_148237394_148237545 | 0.06 |
C7orf33 |
chromosome 7 open reading frame 33 |
50188 |
0.15 |
| chr3_11186255_11186406 | 0.06 |
HRH1 |
histamine receptor H1 |
7551 |
0.29 |
| chr2_65025641_65026023 | 0.06 |
ENSG00000239891 |
. |
18680 |
0.17 |
| chr15_82420642_82420851 | 0.06 |
RP11-597K23.2 |
Uncharacterized protein |
39714 |
0.14 |
| chr10_115753299_115753721 | 0.06 |
ADRB1 |
adrenoceptor beta 1 |
50296 |
0.13 |
| chr4_171593437_171593770 | 0.06 |
ENSG00000251961 |
. |
219015 |
0.02 |
| chr22_37845321_37845613 | 0.06 |
ELFN2 |
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
21962 |
0.12 |
| chr8_104122247_104122563 | 0.06 |
KB-1639H6.4 |
|
10855 |
0.13 |
| chr12_31515195_31515657 | 0.06 |
ENSG00000207477 |
. |
3796 |
0.2 |
| chr7_5676160_5676311 | 0.06 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
41918 |
0.12 |
| chr5_14203788_14204073 | 0.06 |
TRIO |
trio Rho guanine nucleotide exchange factor |
20023 |
0.29 |
| chr3_51655634_51655948 | 0.06 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
7616 |
0.17 |
| chr9_94579747_94580327 | 0.06 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
131125 |
0.05 |
| chr1_82172157_82172358 | 0.06 |
LPHN2 |
latrophilin 2 |
6802 |
0.31 |
| chr5_95630062_95630271 | 0.06 |
ENSG00000206997 |
. |
84235 |
0.09 |
| chr22_46403312_46403647 | 0.06 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
30470 |
0.1 |
| chr8_98992108_98992483 | 0.06 |
MATN2 |
matrilin 2 |
18585 |
0.17 |
| chr15_101656285_101656543 | 0.06 |
RP11-505E24.2 |
|
30143 |
0.19 |
| chr4_116135456_116135745 | 0.06 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
100568 |
0.09 |
| chr1_153493498_153493653 | 0.06 |
BX470102.3 |
|
12504 |
0.08 |
| chr2_44315981_44316132 | 0.06 |
ENSG00000252599 |
. |
65978 |
0.1 |
| chr2_218793055_218793347 | 0.06 |
TNS1 |
tensin 1 |
8269 |
0.24 |
| chr1_152010963_152011114 | 0.06 |
S100A11 |
S100 calcium binding protein A11 |
1527 |
0.33 |
| chr10_90188173_90188378 | 0.06 |
RNLS |
renalase, FAD-dependent amine oxidase |
154672 |
0.04 |
| chr21_40144936_40145370 | 0.06 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
32078 |
0.22 |
| chr2_118981232_118982329 | 0.06 |
INSIG2 |
insulin induced gene 2 |
135730 |
0.05 |
| chr10_80714020_80714184 | 0.06 |
ZMIZ1-AS1 |
ZMIZ1 antisense RNA 1 |
8049 |
0.31 |
| chr3_5051196_5051347 | 0.06 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
29625 |
0.16 |
| chr2_33714938_33715089 | 0.06 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
13297 |
0.26 |
| chr2_65587328_65587537 | 0.06 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
6352 |
0.24 |
| chr7_105793652_105793945 | 0.06 |
SYPL1 |
synaptophysin-like 1 |
40776 |
0.17 |
| chr12_13256087_13256238 | 0.06 |
GSG1 |
germ cell associated 1 |
414 |
0.86 |
| chr14_72409694_72409937 | 0.06 |
RGS6 |
regulator of G-protein signaling 6 |
9866 |
0.26 |
| chr3_119355128_119355463 | 0.06 |
POPDC2 |
popeye domain containing 2 |
24142 |
0.13 |
| chr1_115969093_115969306 | 0.06 |
ENSG00000265534 |
. |
59073 |
0.13 |
| chr3_23780848_23781065 | 0.06 |
ENSG00000238672 |
. |
28146 |
0.18 |
| chr10_108696078_108696229 | 0.06 |
ENSG00000200626 |
. |
33626 |
0.25 |
| chr1_200201358_200201523 | 0.06 |
ENSG00000221403 |
. |
87478 |
0.09 |
| chr4_57157705_57157856 | 0.06 |
ENSG00000252489 |
. |
3030 |
0.25 |
| chr12_65700883_65701407 | 0.06 |
MSRB3 |
methionine sulfoxide reductase B3 |
19510 |
0.22 |
| chr8_59507659_59507955 | 0.06 |
SDCBP |
syndecan binding protein (syntenin) |
30232 |
0.19 |
| chr1_83615938_83616104 | 0.06 |
ENSG00000223231 |
. |
643539 |
0.0 |
| chr8_108395540_108395691 | 0.06 |
ANGPT1 |
angiopoietin 1 |
46865 |
0.2 |
| chr12_94821583_94821926 | 0.06 |
CCDC41 |
coiled-coil domain containing 41 |
31968 |
0.18 |
| chr11_33905413_33905728 | 0.06 |
AC132216.1 |
HCG1785179; PRO1787; Uncharacterized protein |
3381 |
0.25 |
| chr10_49679111_49679316 | 0.06 |
ARHGAP22 |
Rho GTPase activating protein 22 |
19543 |
0.21 |
| chr4_86713761_86713912 | 0.06 |
ARHGAP24 |
Rho GTPase activating protein 24 |
13977 |
0.26 |
| chr17_40244674_40244825 | 0.06 |
DHX58 |
DEXH (Asp-Glu-X-His) box polypeptide 58 |
19732 |
0.09 |
| chr8_25991226_25991377 | 0.06 |
ENSG00000199620 |
. |
3095 |
0.37 |
| chr1_184611707_184611858 | 0.06 |
ENSG00000252790 |
. |
35286 |
0.21 |
| chr10_114817495_114817718 | 0.06 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
86073 |
0.09 |
| chr12_89764903_89765288 | 0.06 |
DUSP6 |
dual specificity phosphatase 6 |
18047 |
0.24 |
| chr16_15687209_15687510 | 0.06 |
CTB-193M12.1 |
|
14972 |
0.15 |
| chr15_71341987_71342266 | 0.06 |
THSD4 |
thrombospondin, type I, domain containing 4 |
47165 |
0.15 |
| chr1_33854701_33854997 | 0.06 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
13655 |
0.15 |
| chr10_77468463_77468644 | 0.06 |
RP11-367B6.2 |
|
33945 |
0.18 |
| chr16_57671396_57671547 | 0.06 |
GPR56 |
G protein-coupled receptor 56 |
1736 |
0.28 |
| chr15_74721212_74721414 | 0.06 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
4688 |
0.16 |
| chr4_8228081_8228232 | 0.06 |
SH3TC1 |
SH3 domain and tetratricopeptide repeats 1 |
10226 |
0.22 |
| chr3_123725960_123726111 | 0.06 |
ROPN1 |
rhophilin associated tail protein 1 |
15010 |
0.2 |
| chr1_204407416_204407567 | 0.06 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
26572 |
0.15 |
| chr10_80176666_80176817 | 0.06 |
ENSG00000201393 |
. |
49477 |
0.19 |
| chr2_165748011_165748162 | 0.06 |
ENSG00000223318 |
. |
4201 |
0.21 |
| chr16_73097967_73098118 | 0.06 |
ZFHX3 |
zinc finger homeobox 3 |
4445 |
0.27 |
| chr16_73221207_73221358 | 0.06 |
C16orf47 |
chromosome 16 open reading frame 47 |
42936 |
0.2 |
| chr10_14782963_14783114 | 0.06 |
FAM107B |
family with sequence similarity 107, member B |
33858 |
0.17 |
| chr21_39856205_39856389 | 0.06 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
14048 |
0.3 |
| chr17_76140274_76140656 | 0.06 |
C17orf99 |
chromosome 17 open reading frame 99 |
1969 |
0.2 |
| chr5_148799600_148799798 | 0.06 |
ENSG00000208035 |
. |
8782 |
0.14 |
| chr1_221334892_221335132 | 0.06 |
HLX |
H2.0-like homeobox |
280428 |
0.01 |
| chr3_194944977_194945345 | 0.06 |
ENSG00000206600 |
. |
9645 |
0.17 |
| chr2_11837137_11837372 | 0.06 |
AC106875.1 |
|
15667 |
0.12 |
| chr7_47497450_47497661 | 0.06 |
TNS3 |
tensin 3 |
4726 |
0.35 |
| chr22_35863792_35863943 | 0.06 |
MCM5 |
minichromosome maintenance complex component 5 |
67443 |
0.1 |
| chr15_101628578_101628729 | 0.06 |
RP11-505E24.2 |
|
2382 |
0.35 |
| chr2_109779392_109779641 | 0.06 |
ENSG00000264934 |
. |
21472 |
0.22 |
| chr8_9018614_9018765 | 0.06 |
RP11-10A14.4 |
Uncharacterized protein |
9376 |
0.17 |
| chr2_152191078_152191290 | 0.06 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
22922 |
0.15 |
| chr12_89555329_89555814 | 0.06 |
ENSG00000238302 |
. |
120491 |
0.06 |
| chr17_13447545_13447833 | 0.06 |
ENSG00000221698 |
. |
726 |
0.71 |
| chr4_74975264_74975487 | 0.06 |
CXCL2 |
chemokine (C-X-C motif) ligand 2 |
10365 |
0.16 |
| chr2_105271366_105271517 | 0.06 |
ENSG00000207249 |
. |
78251 |
0.09 |
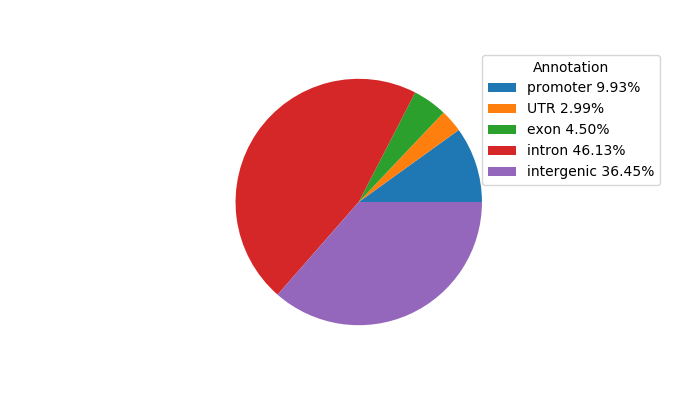
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.1 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.0 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | Genes involved in Phospholipase C-mediated cascade |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |