Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
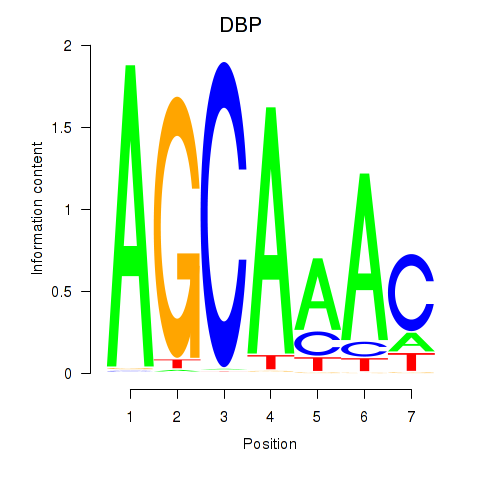
Results for DBP
Z-value: 1.15
Transcription factors associated with DBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBP
|
ENSG00000105516.6 | D-box binding PAR bZIP transcription factor |
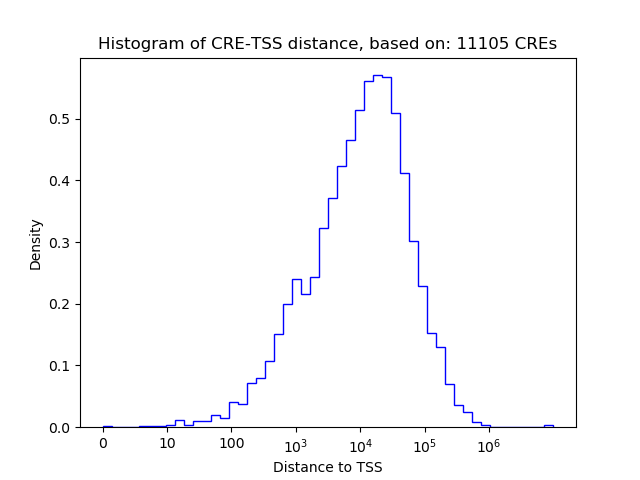
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_49137496_49137669 | DBP | 201 | 0.852337 | 0.64 | 6.3e-02 | Click! |
| chr19_49134368_49134706 | DBP | 3246 | 0.101614 | 0.55 | 1.2e-01 | Click! |
| chr19_49134809_49134960 | DBP | 2899 | 0.108654 | 0.55 | 1.3e-01 | Click! |
| chr19_49133609_49133900 | DBP | 4029 | 0.091434 | 0.34 | 3.7e-01 | Click! |
| chr19_49133360_49133511 | DBP | 4348 | 0.088612 | 0.28 | 4.6e-01 | Click! |
Activity of the DBP motif across conditions
Conditions sorted by the z-value of the DBP motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
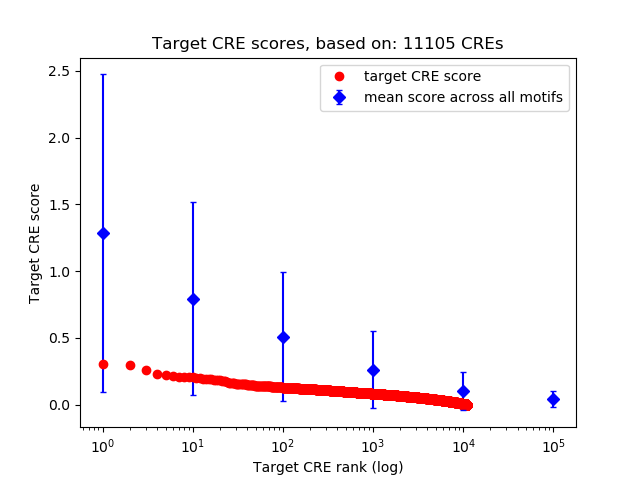
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_46776893_46777044 | 0.30 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
8515 |
0.15 |
| chr2_217339825_217340365 | 0.30 |
AC098820.4 |
|
11824 |
0.15 |
| chr2_136880193_136880455 | 0.26 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
4589 |
0.32 |
| chr1_199012180_199012331 | 0.23 |
ENSG00000239006 |
. |
154057 |
0.04 |
| chr17_34619711_34619862 | 0.22 |
CCL3L1 |
chemokine (C-C motif) ligand 3-like 1 |
5933 |
0.16 |
| chr6_166658110_166658550 | 0.21 |
ENSG00000201292 |
. |
35439 |
0.16 |
| chr17_71663578_71663963 | 0.21 |
SDK2 |
sidekick cell adhesion molecule 2 |
23542 |
0.21 |
| chr17_33694633_33695079 | 0.21 |
SLFN11 |
schlafen family member 11 |
5770 |
0.2 |
| chr10_23828998_23829406 | 0.21 |
OTUD1 |
OTU domain containing 1 |
101004 |
0.07 |
| chr3_128718043_128718322 | 0.21 |
EFCC1 |
EF-hand and coiled-coil domain containing 1 |
2290 |
0.23 |
| chr17_34518155_34518317 | 0.20 |
AC131056.3 |
|
2908 |
0.15 |
| chr17_34606209_34606509 | 0.20 |
TBC1D3C |
TBC1 domain family, member 3C |
14327 |
0.13 |
| chr17_34411882_34412121 | 0.20 |
AC069363.1 |
|
3009 |
0.13 |
| chr22_32538380_32538635 | 0.20 |
AP1B1P1 |
adaptor-related protein complex 1, beta 1 subunit pseudogene 1 |
10203 |
0.14 |
| chr10_105672200_105672613 | 0.19 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
5021 |
0.2 |
| chr12_9916868_9917340 | 0.19 |
CD69 |
CD69 molecule |
3607 |
0.2 |
| chr4_103656767_103656918 | 0.19 |
MANBA |
mannosidase, beta A, lysosomal |
25248 |
0.2 |
| chr12_64994089_64994240 | 0.19 |
RP11-338E21.1 |
|
9075 |
0.13 |
| chr15_55539751_55540118 | 0.18 |
RAB27A |
RAB27A, member RAS oncogene family |
1299 |
0.5 |
| chr18_74835056_74835207 | 0.18 |
MBP |
myelin basic protein |
4537 |
0.34 |
| chr4_79535031_79535295 | 0.18 |
ENSG00000200998 |
. |
18376 |
0.2 |
| chr6_147091242_147091431 | 0.18 |
ADGB |
androglobin |
239 |
0.96 |
| chr9_92123694_92124049 | 0.18 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
10826 |
0.22 |
| chr17_47838646_47838824 | 0.17 |
FAM117A |
family with sequence similarity 117, member A |
2758 |
0.22 |
| chr14_34649904_34650139 | 0.16 |
EGLN3 |
egl-9 family hypoxia-inducible factor 3 |
42062 |
0.2 |
| chr2_234318940_234319134 | 0.16 |
DGKD |
diacylglycerol kinase, delta 130kDa |
22237 |
0.15 |
| chr11_118096084_118096517 | 0.16 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
491 |
0.74 |
| chr2_157327257_157327445 | 0.16 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
2733 |
0.29 |
| chr8_102212410_102212571 | 0.16 |
ZNF706 |
zinc finger protein 706 |
1395 |
0.52 |
| chr19_16704377_16704738 | 0.16 |
CTD-3222D19.5 |
|
5761 |
0.09 |
| chr7_50356212_50356462 | 0.16 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
8019 |
0.29 |
| chr6_166849014_166849195 | 0.16 |
RPS6KA2-IT1 |
RPS6KA2 intronic transcript 1 (non-protein coding) |
29767 |
0.15 |
| chr4_8222732_8223119 | 0.16 |
SH3TC1 |
SH3 domain and tetratricopeptide repeats 1 |
4995 |
0.25 |
| chr2_158349915_158350402 | 0.16 |
CYTIP |
cytohesin 1 interacting protein |
4685 |
0.29 |
| chr2_54817252_54817551 | 0.16 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
31870 |
0.16 |
| chr7_12262250_12262487 | 0.16 |
TMEM106B |
transmembrane protein 106B |
11419 |
0.31 |
| chr16_50719380_50719617 | 0.16 |
SNX20 |
sorting nexin 20 |
4234 |
0.14 |
| chr1_200098445_200098816 | 0.16 |
ENSG00000221403 |
. |
15332 |
0.24 |
| chr9_74245405_74245556 | 0.15 |
TMEM2 |
transmembrane protein 2 |
74368 |
0.12 |
| chr6_6658591_6658742 | 0.15 |
LY86-AS1 |
LY86 antisense RNA 1 |
35662 |
0.21 |
| chr5_122227538_122227689 | 0.15 |
SNX24 |
sorting nexin 24 |
46292 |
0.13 |
| chr10_70846687_70847039 | 0.15 |
SRGN |
serglycin |
1011 |
0.55 |
| chr10_48478469_48478908 | 0.15 |
GDF10 |
growth differentiation factor 10 |
39712 |
0.15 |
| chr20_11872259_11872563 | 0.15 |
BTBD3 |
BTB (POZ) domain containing 3 |
436 |
0.88 |
| chr1_167596920_167597366 | 0.15 |
RCSD1 |
RCSD domain containing 1 |
2187 |
0.29 |
| chr14_89924951_89925223 | 0.15 |
FOXN3 |
forkhead box N3 |
35308 |
0.14 |
| chr7_35076257_35076437 | 0.15 |
DPY19L1 |
dpy-19-like 1 (C. elegans) |
1306 |
0.58 |
| chr11_128571263_128571500 | 0.15 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
5463 |
0.2 |
| chr18_74716586_74716828 | 0.14 |
MBP |
myelin basic protein |
5371 |
0.27 |
| chr19_16228666_16229162 | 0.14 |
RAB8A |
RAB8A, member RAS oncogene family |
6207 |
0.14 |
| chr17_8859855_8860380 | 0.14 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
8907 |
0.24 |
| chr2_39444415_39444721 | 0.14 |
CDKL4 |
cyclin-dependent kinase-like 4 |
12105 |
0.21 |
| chr19_2281233_2281423 | 0.14 |
C19orf35 |
chromosome 19 open reading frame 35 |
847 |
0.38 |
| chr18_56382154_56382522 | 0.14 |
RP11-126O1.4 |
|
37492 |
0.12 |
| chr14_20955809_20956004 | 0.14 |
PNP |
purine nucleoside phosphorylase |
17221 |
0.08 |
| chr17_7238561_7238964 | 0.14 |
ACAP1 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
1086 |
0.22 |
| chr1_31231275_31231444 | 0.14 |
LAPTM5 |
lysosomal protein transmembrane 5 |
692 |
0.66 |
| chr13_31784399_31784550 | 0.14 |
B3GALTL |
beta 1,3-galactosyltransferase-like |
10401 |
0.27 |
| chr4_55811439_55811670 | 0.14 |
ENSG00000264332 |
. |
65545 |
0.11 |
| chr5_115695176_115695329 | 0.14 |
CTB-118N6.3 |
|
87991 |
0.09 |
| chr6_6611197_6611375 | 0.14 |
LY86-AS1 |
LY86 antisense RNA 1 |
11642 |
0.25 |
| chr9_136208045_136208226 | 0.14 |
ENSG00000201451 |
. |
3474 |
0.08 |
| chr22_17588681_17588894 | 0.14 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
13356 |
0.14 |
| chr3_46342500_46342931 | 0.14 |
CCR3 |
chemokine (C-C motif) receptor 3 |
36096 |
0.14 |
| chr17_2695452_2695668 | 0.14 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
4172 |
0.2 |
| chr7_50353474_50353683 | 0.14 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
5260 |
0.32 |
| chr21_43688719_43689075 | 0.14 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
12438 |
0.16 |
| chr18_52502942_52503093 | 0.14 |
RAB27B |
RAB27B, member RAS oncogene family |
7587 |
0.27 |
| chr15_94802319_94802604 | 0.14 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
27694 |
0.27 |
| chr1_150975197_150975452 | 0.14 |
FAM63A |
family with sequence similarity 63, member A |
3637 |
0.1 |
| chr16_68109101_68109477 | 0.14 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
9958 |
0.1 |
| chr22_50838176_50838422 | 0.14 |
ENSG00000238604 |
. |
1050 |
0.38 |
| chr5_150591857_150592074 | 0.14 |
GM2A |
GM2 ganglioside activator |
254 |
0.92 |
| chr15_31515349_31515542 | 0.14 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
61969 |
0.13 |
| chr13_32609571_32609811 | 0.14 |
FRY-AS1 |
FRY antisense RNA 1 |
3915 |
0.26 |
| chr15_88304946_88305195 | 0.13 |
NTRK3 |
neurotrophic tyrosine kinase, receptor, type 3 |
171251 |
0.04 |
| chr5_107008246_107008397 | 0.13 |
EFNA5 |
ephrin-A5 |
1725 |
0.5 |
| chr2_233924263_233924811 | 0.13 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
140 |
0.96 |
| chr16_46692550_46692750 | 0.13 |
RP11-93O14.2 |
|
2853 |
0.24 |
| chr12_82343405_82343568 | 0.13 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
190154 |
0.03 |
| chr7_8216722_8216994 | 0.13 |
ENSG00000265212 |
. |
22206 |
0.19 |
| chr9_117701644_117701992 | 0.13 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
9121 |
0.28 |
| chr12_11824151_11824362 | 0.13 |
ETV6 |
ets variant 6 |
21468 |
0.25 |
| chr7_37349133_37349657 | 0.13 |
ELMO1 |
engulfment and cell motility 1 |
32972 |
0.18 |
| chr12_66661258_66661535 | 0.13 |
RP11-335I12.2 |
|
10182 |
0.16 |
| chr4_68441371_68441728 | 0.13 |
STAP1 |
signal transducing adaptor family member 1 |
17103 |
0.2 |
| chr2_111562957_111563362 | 0.13 |
ACOXL |
acyl-CoA oxidase-like |
263 |
0.95 |
| chr18_60766372_60766612 | 0.13 |
RP11-299P2.1 |
|
52061 |
0.15 |
| chr6_108145921_108146601 | 0.13 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
740 |
0.76 |
| chr17_17659846_17659997 | 0.13 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
14214 |
0.13 |
| chr5_169706707_169706999 | 0.13 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
12522 |
0.22 |
| chr13_42036238_42036522 | 0.13 |
RGCC |
regulator of cell cycle |
4685 |
0.18 |
| chr3_18766132_18766453 | 0.13 |
ENSG00000228956 |
. |
20944 |
0.29 |
| chr12_102171783_102172089 | 0.13 |
ENSG00000201168 |
. |
1656 |
0.23 |
| chr7_657809_658035 | 0.13 |
AC147651.4 |
|
15100 |
0.17 |
| chr1_28196447_28196641 | 0.13 |
THEMIS2 |
thymocyte selection associated family member 2 |
2511 |
0.17 |
| chr6_139565905_139566056 | 0.13 |
RP1-225E12.2 |
|
5538 |
0.21 |
| chr1_40503504_40503714 | 0.13 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
2296 |
0.3 |
| chr10_98411356_98411597 | 0.13 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
17892 |
0.19 |
| chr8_142131719_142131931 | 0.13 |
DENND3 |
DENN/MADD domain containing 3 |
4448 |
0.23 |
| chr7_8216150_8216370 | 0.13 |
ENSG00000265212 |
. |
21608 |
0.19 |
| chr18_29621140_29621437 | 0.13 |
ENSG00000265063 |
. |
19664 |
0.13 |
| chr1_229162847_229163204 | 0.13 |
RP5-1061H20.5 |
|
200284 |
0.02 |
| chr8_134562977_134563358 | 0.13 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
20925 |
0.27 |
| chr14_90185242_90185495 | 0.13 |
ENSG00000200312 |
. |
6513 |
0.24 |
| chrX_128917643_128917877 | 0.13 |
SASH3 |
SAM and SH3 domain containing 3 |
3800 |
0.24 |
| chr1_159868738_159869013 | 0.13 |
CCDC19 |
coiled-coil domain containing 19 |
816 |
0.34 |
| chr20_20664895_20665109 | 0.13 |
RALGAPA2 |
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
28129 |
0.23 |
| chr15_70780606_70780757 | 0.13 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
213939 |
0.02 |
| chr9_115627506_115627657 | 0.13 |
RP11-408O19.5 |
|
19857 |
0.17 |
| chr2_68652249_68652409 | 0.13 |
FBXO48 |
F-box protein 48 |
42061 |
0.12 |
| chr7_37389453_37389714 | 0.13 |
ENSG00000222869 |
. |
1164 |
0.51 |
| chrX_65267830_65268055 | 0.13 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
7975 |
0.23 |
| chr15_31271292_31271496 | 0.13 |
MTMR10 |
myotubularin related protein 10 |
4802 |
0.18 |
| chr10_7526915_7527101 | 0.13 |
ENSG00000207453 |
. |
1763 |
0.44 |
| chr2_136899107_136899528 | 0.13 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
23582 |
0.25 |
| chr12_8755911_8756062 | 0.13 |
AICDA |
activation-induced cytidine deaminase |
3626 |
0.16 |
| chr1_207232696_207232912 | 0.13 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
4759 |
0.14 |
| chr11_64976530_64976899 | 0.13 |
CAPN1 |
calpain 1, (mu/I) large subunit |
25907 |
0.08 |
| chr15_101071565_101071716 | 0.13 |
CERS3 |
ceramide synthase 3 |
12836 |
0.15 |
| chr5_169306526_169306677 | 0.13 |
CTB-37A13.1 |
|
100232 |
0.07 |
| chr8_142414415_142414628 | 0.13 |
CTD-3064M3.4 |
|
11847 |
0.12 |
| chr13_46743886_46744372 | 0.13 |
ENSG00000240767 |
. |
246 |
0.88 |
| chr1_3807311_3807510 | 0.13 |
ENSG00000264428 |
. |
6782 |
0.16 |
| chr10_14348378_14348529 | 0.13 |
FRMD4A |
FERM domain containing 4A |
23648 |
0.23 |
| chr20_8310366_8310517 | 0.12 |
PLCB1-IT1 |
PLCB1 intronic transcript 1 (non-protein coding) |
81069 |
0.11 |
| chr17_47824587_47824834 | 0.12 |
FAM117A |
family with sequence similarity 117, member A |
16783 |
0.14 |
| chr15_86251203_86251443 | 0.12 |
RP11-815J21.1 |
|
6983 |
0.12 |
| chr15_63740013_63740164 | 0.12 |
USP3 |
ubiquitin specific peptidase 3 |
56705 |
0.13 |
| chr18_52384635_52384905 | 0.12 |
RAB27B |
RAB27B, member RAS oncogene family |
321 |
0.94 |
| chr1_221884647_221884930 | 0.12 |
DUSP10 |
dual specificity phosphatase 10 |
26014 |
0.26 |
| chr1_25361172_25361323 | 0.12 |
ENSG00000264371 |
. |
11253 |
0.23 |
| chr8_125046706_125046857 | 0.12 |
RP11-959I15.4 |
|
5928 |
0.2 |
| chr2_8562176_8562384 | 0.12 |
AC011747.7 |
|
253616 |
0.02 |
| chrX_128904932_128905357 | 0.12 |
SASH3 |
SAM and SH3 domain containing 3 |
8816 |
0.2 |
| chr3_112183882_112184126 | 0.12 |
BTLA |
B and T lymphocyte associated |
34201 |
0.19 |
| chr12_111065801_111066057 | 0.12 |
ENSG00000263537 |
. |
2207 |
0.23 |
| chr19_2619378_2619607 | 0.12 |
CTC-265F19.2 |
|
7632 |
0.14 |
| chr22_38068883_38069075 | 0.12 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
2636 |
0.13 |
| chr11_128607706_128608101 | 0.12 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
26782 |
0.17 |
| chr15_63898445_63898817 | 0.12 |
USP3-AS1 |
USP3 antisense RNA 1 |
4843 |
0.21 |
| chr8_37731564_37731977 | 0.12 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
3865 |
0.18 |
| chr20_57829512_57829876 | 0.12 |
EDN3 |
endothelin 3 |
45788 |
0.15 |
| chr14_91845514_91845690 | 0.12 |
CCDC88C |
coiled-coil domain containing 88C |
38088 |
0.16 |
| chr20_19314143_19314471 | 0.12 |
ENSG00000221748 |
. |
7633 |
0.26 |
| chr11_33255190_33255341 | 0.12 |
HIPK3 |
homeodomain interacting protein kinase 3 |
22953 |
0.17 |
| chr6_91060753_91061282 | 0.12 |
ENSG00000266760 |
. |
38556 |
0.18 |
| chr17_45781212_45781384 | 0.12 |
TBKBP1 |
TBK1 binding protein 1 |
8668 |
0.14 |
| chr2_64695090_64695299 | 0.12 |
LGALSL |
lectin, galactoside-binding-like |
13518 |
0.19 |
| chr3_12503836_12504037 | 0.12 |
TSEN2 |
TSEN2 tRNA splicing endonuclease subunit |
21995 |
0.15 |
| chr13_60026787_60027144 | 0.12 |
ENSG00000239003 |
. |
27239 |
0.28 |
| chr16_50743382_50743603 | 0.12 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
2613 |
0.16 |
| chr10_73535863_73536136 | 0.12 |
C10orf54 |
chromosome 10 open reading frame 54 |
2744 |
0.26 |
| chr21_47788798_47788949 | 0.12 |
PCNT |
pericentrin |
44837 |
0.1 |
| chr22_17578526_17578936 | 0.12 |
IL17RA |
interleukin 17 receptor A |
12882 |
0.15 |
| chr3_195277399_195277835 | 0.12 |
AC091633.3 |
|
4198 |
0.18 |
| chr1_108586650_108587048 | 0.12 |
ENSG00000264753 |
. |
25456 |
0.21 |
| chrX_138636733_138636884 | 0.12 |
F9 |
coagulation factor IX |
23884 |
0.2 |
| chr1_26616916_26617250 | 0.12 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
10470 |
0.11 |
| chr10_26667989_26668241 | 0.12 |
APBB1IP |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
59017 |
0.15 |
| chr1_154376165_154376624 | 0.12 |
IL6R |
interleukin 6 receptor |
1275 |
0.33 |
| chr17_72726639_72727007 | 0.12 |
RAB37 |
RAB37, member RAS oncogene family |
5914 |
0.11 |
| chr10_6607286_6607516 | 0.12 |
PRKCQ |
protein kinase C, theta |
14800 |
0.3 |
| chr1_92925228_92925479 | 0.12 |
GFI1 |
growth factor independent 1 transcription repressor |
24158 |
0.21 |
| chr12_69742182_69742355 | 0.12 |
LYZ |
lysozyme |
102 |
0.96 |
| chr1_40308954_40309106 | 0.12 |
ENSG00000202222 |
. |
38320 |
0.09 |
| chr1_36922243_36922595 | 0.12 |
OSCP1 |
organic solute carrier partner 1 |
6333 |
0.14 |
| chr2_136893672_136893831 | 0.12 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
18016 |
0.27 |
| chr7_38349303_38349454 | 0.12 |
STARD3NL |
STARD3 N-terminal like |
131381 |
0.05 |
| chr5_118643731_118644037 | 0.12 |
ENSG00000243333 |
. |
1558 |
0.41 |
| chr2_148806923_148807074 | 0.12 |
ORC4 |
origin recognition complex, subunit 4 |
27851 |
0.19 |
| chr2_240162756_240163017 | 0.12 |
HDAC4 |
histone deacetylase 4 |
50134 |
0.11 |
| chrX_70391376_70391679 | 0.12 |
RP5-1091N2.9 |
|
26498 |
0.1 |
| chr5_86375411_86375667 | 0.12 |
ENSG00000251951 |
. |
23124 |
0.21 |
| chr7_157182751_157183382 | 0.12 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
50551 |
0.15 |
| chr2_74268024_74268228 | 0.12 |
TET3 |
tet methylcytosine dioxygenase 3 |
5324 |
0.2 |
| chr5_40670687_40670879 | 0.12 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
8817 |
0.21 |
| chr3_30644339_30644515 | 0.12 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
3567 |
0.37 |
| chr12_29362316_29362582 | 0.12 |
FAR2 |
fatty acyl CoA reductase 2 |
14149 |
0.26 |
| chr8_119121685_119121918 | 0.12 |
EXT1 |
exostosin glycosyltransferase 1 |
852 |
0.77 |
| chr11_121340734_121341098 | 0.12 |
RP11-730K11.1 |
|
17194 |
0.23 |
| chr2_70667703_70667865 | 0.12 |
ENSG00000215996 |
. |
14972 |
0.15 |
| chr11_48044517_48044998 | 0.12 |
AC103828.1 |
|
7350 |
0.22 |
| chr22_19084512_19084678 | 0.12 |
AC004471.9 |
|
24447 |
0.1 |
| chr15_70007265_70007861 | 0.12 |
ENSG00000238870 |
. |
15598 |
0.26 |
| chr7_96467239_96467390 | 0.12 |
ENSG00000244318 |
. |
102362 |
0.07 |
| chr17_55358360_55358511 | 0.12 |
MSI2 |
musashi RNA-binding protein 2 |
4770 |
0.31 |
| chr20_45181455_45181613 | 0.12 |
OCSTAMP |
osteoclast stimulatory transmembrane protein |
2321 |
0.32 |
| chr7_114776119_114776270 | 0.12 |
ENSG00000199764 |
. |
71856 |
0.13 |
| chr3_16887733_16887892 | 0.12 |
PLCL2 |
phospholipase C-like 2 |
38640 |
0.2 |
| chr14_89819638_89820075 | 0.12 |
RP11-356K23.2 |
|
1548 |
0.36 |
| chr2_8007405_8007668 | 0.12 |
ENSG00000221255 |
. |
290564 |
0.01 |
| chr17_28275822_28275973 | 0.12 |
EFCAB5 |
EF-hand calcium binding domain 5 |
7274 |
0.16 |
| chr7_130056827_130057089 | 0.12 |
CEP41 |
centrosomal protein 41kDa |
215 |
0.92 |
| chr15_92401739_92402176 | 0.12 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
4606 |
0.29 |
| chr6_24950964_24951248 | 0.12 |
FAM65B |
family with sequence similarity 65, member B |
14918 |
0.22 |
| chr8_130600986_130601137 | 0.12 |
ENSG00000266387 |
. |
91148 |
0.06 |
| chr20_4792096_4792247 | 0.12 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
3598 |
0.25 |
| chr7_139314548_139314773 | 0.12 |
CLEC2L |
C-type lectin domain family 2, member L |
105832 |
0.06 |
| chr13_33562441_33562592 | 0.12 |
KL |
klotho |
27691 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.2 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.1 | 0.2 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.1 | 0.3 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.0 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.1 | GO:0046469 | platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0032616 | interleukin-13 production(GO:0032616) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0033084 | regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0010829 | negative regulation of glucose transport(GO:0010829) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0002883 | hypersensitivity(GO:0002524) regulation of hypersensitivity(GO:0002883) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0090181 | regulation of cholesterol metabolic process(GO:0090181) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.0 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.0 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.1 | GO:0072505 | phosphate ion homeostasis(GO:0055062) divalent inorganic anion homeostasis(GO:0072505) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.0 | 0.1 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.0 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.1 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.2 | GO:0070664 | negative regulation of leukocyte proliferation(GO:0070664) |
| 0.0 | 0.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.5 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.0 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0000036 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.4 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.0 | GO:0004556 | alpha-amylase activity(GO:0004556) amylase activity(GO:0016160) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.0 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.0 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.0 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |