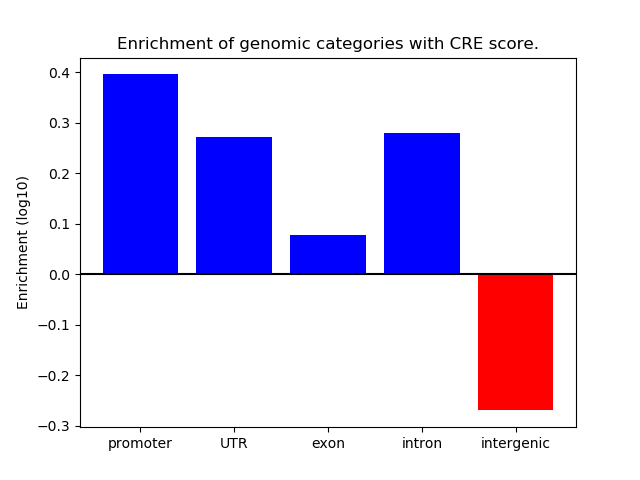
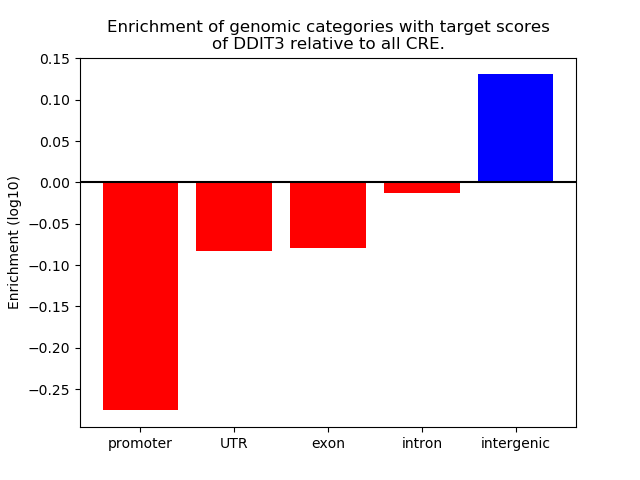
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
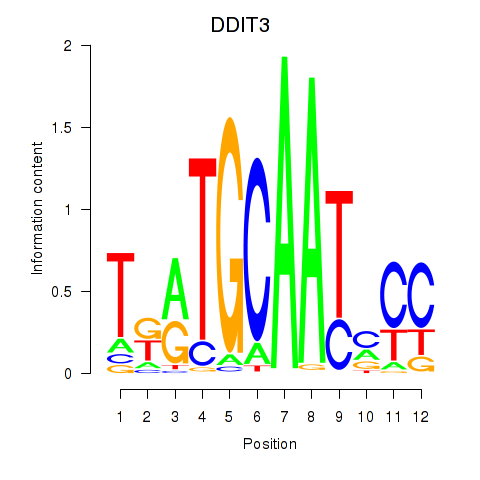
Results for DDIT3
Z-value: 1.77
Transcription factors associated with DDIT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DDIT3
|
ENSG00000175197.6 | DNA damage inducible transcript 3 |
Activity of the DDIT3 motif across conditions
Conditions sorted by the z-value of the DDIT3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
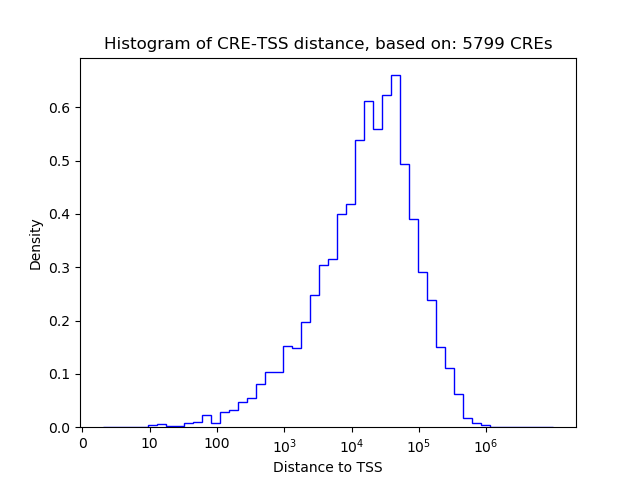
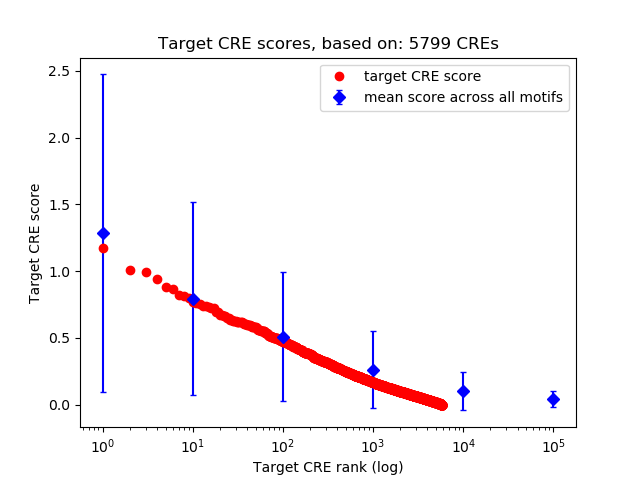
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_66289441_66289878 | 1.17 |
RP11-366L20.2 |
Uncharacterized protein |
13712 |
0.17 |
| chr9_84106067_84106472 | 1.01 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
122168 |
0.06 |
| chr20_23238843_23239098 | 0.99 |
NXT1 |
NTF2-like export factor 1 |
92403 |
0.05 |
| chr11_43529872_43530198 | 0.94 |
ENSG00000211568 |
. |
51171 |
0.12 |
| chr2_145402982_145403264 | 0.88 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
124502 |
0.06 |
| chr8_62630791_62631038 | 0.87 |
ENSG00000264408 |
. |
3567 |
0.27 |
| chr6_18216555_18216818 | 0.82 |
KDM1B |
lysine (K)-specific demethylase 1B |
30667 |
0.15 |
| chr7_17408913_17409156 | 0.81 |
ENSG00000199473 |
. |
3916 |
0.32 |
| chr2_113998730_113999135 | 0.80 |
PAX8 |
paired box 8 |
328 |
0.68 |
| chr2_127934430_127934663 | 0.77 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
28797 |
0.2 |
| chr10_93884742_93884893 | 0.76 |
ENSG00000264076 |
. |
34512 |
0.16 |
| chr2_12959099_12960371 | 0.76 |
ENSG00000264370 |
. |
82242 |
0.11 |
| chr5_77889982_77890133 | 0.74 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
45083 |
0.2 |
| chr13_42270469_42270620 | 0.74 |
ENSG00000241406 |
. |
111269 |
0.06 |
| chr1_232840317_232840703 | 0.73 |
ENSG00000238382 |
. |
4560 |
0.32 |
| chr3_30333675_30334003 | 0.72 |
ENSG00000199927 |
. |
12188 |
0.3 |
| chr7_77082891_77083367 | 0.72 |
ENSG00000238453 |
. |
30024 |
0.19 |
| chr20_30198397_30198778 | 0.70 |
ENSG00000264395 |
. |
3598 |
0.15 |
| chr11_34586202_34586499 | 0.69 |
ELF5 |
E74-like factor 5 (ets domain transcription factor) |
50998 |
0.14 |
| chr4_170187898_170188049 | 0.68 |
SH3RF1 |
SH3 domain containing ring finger 1 |
3135 |
0.35 |
| chr21_44141550_44141701 | 0.67 |
AP001627.1 |
|
20243 |
0.2 |
| chr10_95193664_95193870 | 0.67 |
MYOF |
myoferlin |
48184 |
0.14 |
| chr4_13921909_13922060 | 0.67 |
ENSG00000252092 |
. |
261733 |
0.02 |
| chr2_26981199_26981390 | 0.65 |
SLC35F6 |
solute carrier family 35, member F6 |
5862 |
0.19 |
| chr15_73979466_73979617 | 0.65 |
CD276 |
CD276 molecule |
2194 |
0.36 |
| chr3_128914988_128915202 | 0.64 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
12330 |
0.14 |
| chr16_88058565_88058905 | 0.64 |
BANP |
BTG3 associated nuclear protein |
55111 |
0.14 |
| chr1_42083758_42083983 | 0.63 |
RP11-486B10.3 |
|
82756 |
0.09 |
| chr14_51404344_51404736 | 0.62 |
PYGL |
phosphorylase, glycogen, liver |
6634 |
0.19 |
| chr2_26232303_26232586 | 0.62 |
AC013449.1 |
Uncharacterized protein |
19037 |
0.15 |
| chr2_162969937_162970139 | 0.62 |
GCG |
glucagon |
38875 |
0.13 |
| chr3_134050665_134050816 | 0.62 |
AMOTL2 |
angiomotin like 2 |
40014 |
0.16 |
| chr2_146996340_146996491 | 0.62 |
ENSG00000251905 |
. |
93691 |
0.1 |
| chr10_74084780_74085101 | 0.62 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
19779 |
0.16 |
| chr13_106770061_106770212 | 0.62 |
ENSG00000222643 |
. |
10584 |
0.27 |
| chr8_116461452_116461961 | 0.61 |
TRPS1 |
trichorhinophalangeal syndrome I |
42742 |
0.19 |
| chr11_58733011_58733162 | 0.61 |
RP11-142C4.6 |
|
13672 |
0.16 |
| chr3_133289241_133289476 | 0.61 |
CDV3 |
CDV3 homolog (mouse) |
3216 |
0.26 |
| chr4_86953575_86953726 | 0.60 |
RP13-514E23.1 |
|
18623 |
0.19 |
| chr13_45389687_45389838 | 0.60 |
NUFIP1 |
nuclear fragile X mental retardation protein interacting protein 1 |
173856 |
0.03 |
| chr5_34487322_34487473 | 0.60 |
RAI14 |
retinoic acid induced 14 |
168945 |
0.03 |
| chr3_14246546_14246821 | 0.60 |
XPC |
xeroderma pigmentosum, complementation group C |
26400 |
0.16 |
| chr7_16697056_16697503 | 0.59 |
BZW2 |
basic leucine zipper and W2 domains 2 |
3569 |
0.24 |
| chr1_56981658_56981809 | 0.59 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
63508 |
0.14 |
| chr1_201681170_201681321 | 0.59 |
ENSG00000264802 |
. |
7391 |
0.14 |
| chr2_216765021_216765281 | 0.58 |
ENSG00000212055 |
. |
21509 |
0.25 |
| chr20_10286775_10287859 | 0.58 |
ENSG00000211588 |
. |
55561 |
0.13 |
| chr7_38784313_38784750 | 0.58 |
VPS41 |
vacuolar protein sorting 41 homolog (S. cerevisiae) |
631 |
0.81 |
| chr6_155815833_155816170 | 0.58 |
NOX3 |
NADPH oxidase 3 |
38964 |
0.21 |
| chr1_86038271_86038452 | 0.58 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
5572 |
0.2 |
| chr5_151043511_151043918 | 0.58 |
CTB-113P19.4 |
|
11878 |
0.14 |
| chr2_40544107_40544258 | 0.57 |
AC007377.1 |
|
56035 |
0.15 |
| chr11_57045647_57045894 | 0.56 |
APLNR |
apelin receptor |
41095 |
0.09 |
| chr1_56278160_56278311 | 0.56 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
126718 |
0.06 |
| chr1_204381057_204381208 | 0.56 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
213 |
0.94 |
| chr9_72619163_72619314 | 0.56 |
MAMDC2 |
MAM domain containing 2 |
39259 |
0.18 |
| chr2_9374494_9374810 | 0.55 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
27758 |
0.24 |
| chr4_157672395_157672563 | 0.55 |
RP11-154F14.2 |
|
90032 |
0.08 |
| chr15_63232625_63232776 | 0.55 |
RP11-1069G10.1 |
|
56467 |
0.12 |
| chr6_138231300_138231872 | 0.55 |
RP11-356I2.4 |
|
42216 |
0.16 |
| chr17_43407056_43407601 | 0.55 |
ENSG00000199953 |
. |
2465 |
0.22 |
| chr12_52077625_52077776 | 0.55 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
2507 |
0.37 |
| chr7_137627516_137627793 | 0.55 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
9018 |
0.21 |
| chr6_7975682_7975960 | 0.54 |
TXNDC5 |
thioredoxin domain containing 5 (endoplasmic reticulum) |
50825 |
0.16 |
| chr14_85983042_85983193 | 0.54 |
RP11-497E19.2 |
Uncharacterized protein |
11826 |
0.25 |
| chr5_169002894_169003181 | 0.53 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
7601 |
0.23 |
| chr5_37771599_37771776 | 0.53 |
ENSG00000238335 |
. |
30480 |
0.2 |
| chr8_129071045_129071196 | 0.53 |
ENSG00000221176 |
. |
9722 |
0.21 |
| chr11_58394708_58394859 | 0.53 |
CNTF |
ciliary neurotrophic factor |
4637 |
0.18 |
| chr3_154736890_154737041 | 0.52 |
MME |
membrane metallo-endopeptidase |
4948 |
0.35 |
| chrX_68357539_68357690 | 0.52 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
24923 |
0.27 |
| chr5_65543145_65543296 | 0.52 |
ENSG00000238400 |
. |
16160 |
0.26 |
| chr3_171528829_171529427 | 0.51 |
PLD1 |
phospholipase D1, phosphatidylcholine-specific |
844 |
0.52 |
| chr21_43518310_43518552 | 0.51 |
C21orf128 |
chromosome 21 open reading frame 128 |
10213 |
0.19 |
| chr5_90421803_90421954 | 0.51 |
ENSG00000199643 |
. |
144666 |
0.05 |
| chr10_128947522_128947673 | 0.51 |
FAM196A |
family with sequence similarity 196, member A |
27676 |
0.24 |
| chr6_13762651_13763628 | 0.51 |
MCUR1 |
mitochondrial calcium uniporter regulator 1 |
38438 |
0.15 |
| chr4_39152584_39152984 | 0.50 |
RP11-360F5.1 |
|
24346 |
0.17 |
| chr10_116755757_116755908 | 0.50 |
ENSG00000252611 |
. |
26505 |
0.22 |
| chr1_64211693_64212078 | 0.50 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
27808 |
0.21 |
| chr5_64496427_64496578 | 0.50 |
ENSG00000207439 |
. |
77306 |
0.12 |
| chr8_22811234_22811445 | 0.50 |
ENSG00000240116 |
. |
24051 |
0.12 |
| chr12_72104450_72104601 | 0.50 |
TMEM19 |
transmembrane protein 19 |
13293 |
0.18 |
| chr3_114326741_114326892 | 0.50 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
16237 |
0.28 |
| chr1_201595609_201595975 | 0.50 |
NAV1 |
neuron navigator 1 |
3191 |
0.21 |
| chr13_76290473_76290624 | 0.50 |
LMO7 |
LIM domain 7 |
43955 |
0.16 |
| chr1_155618391_155618704 | 0.49 |
YY1AP1 |
YY1 associated protein 1 |
27915 |
0.11 |
| chr15_42222648_42223001 | 0.49 |
CTD-2382E5.4 |
|
9209 |
0.12 |
| chr14_95130458_95130609 | 0.49 |
SERPINA13P |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 13, pseudogene |
22878 |
0.19 |
| chr1_11218024_11218175 | 0.49 |
ENSG00000253086 |
. |
5692 |
0.12 |
| chr9_108004052_108004203 | 0.49 |
SLC44A1 |
solute carrier family 44 (choline transporter), member 1 |
2776 |
0.4 |
| chr10_81161241_81161392 | 0.49 |
RP11-342M3.5 |
|
19232 |
0.18 |
| chr3_188014840_188014991 | 0.48 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
57263 |
0.15 |
| chr11_122577004_122577557 | 0.48 |
ENSG00000239079 |
. |
19739 |
0.22 |
| chr4_143345126_143345417 | 0.48 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
7141 |
0.35 |
| chr10_17041586_17041737 | 0.48 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
15467 |
0.27 |
| chr11_86658361_86658657 | 0.48 |
FZD4 |
frizzled family receptor 4 |
7924 |
0.24 |
| chr13_34527970_34528121 | 0.47 |
ENSG00000263663 |
. |
41916 |
0.2 |
| chr11_112031799_112032015 | 0.47 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
2890 |
0.14 |
| chr14_52847171_52847322 | 0.47 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
66133 |
0.12 |
| chr9_111274185_111274336 | 0.47 |
ENSG00000222512 |
. |
153051 |
0.04 |
| chr13_110746918_110747075 | 0.47 |
ENSG00000265885 |
. |
3502 |
0.37 |
| chr6_155384408_155384559 | 0.47 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
26940 |
0.24 |
| chr5_67192624_67192790 | 0.47 |
ENSG00000223149 |
. |
70566 |
0.14 |
| chr17_62843312_62843537 | 0.47 |
PLEKHM1P |
pleckstrin homology domain containing, family M (with RUN domain) member 1 pseudogene |
10181 |
0.12 |
| chr1_201728441_201728592 | 0.47 |
NAV1 |
neuron navigator 1 |
19520 |
0.11 |
| chr6_11207055_11207535 | 0.47 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
25596 |
0.18 |
| chr7_130934065_130934342 | 0.47 |
MKLN1 |
muskelin 1, intracellular mediator containing kelch motifs |
35618 |
0.2 |
| chr1_181472407_181472558 | 0.47 |
CACNA1E |
calcium channel, voltage-dependent, R type, alpha 1E subunit |
19601 |
0.29 |
| chr10_54207743_54207894 | 0.46 |
RP11-556E13.1 |
|
113047 |
0.07 |
| chr1_101616339_101616490 | 0.46 |
ENSG00000252765 |
. |
17705 |
0.19 |
| chr11_128778051_128778202 | 0.46 |
C11orf45 |
chromosome 11 open reading frame 45 |
2196 |
0.25 |
| chr17_8762469_8762816 | 0.46 |
PIK3R6 |
phosphoinositide-3-kinase, regulatory subunit 6 |
8352 |
0.2 |
| chr11_125491960_125492111 | 0.46 |
CHEK1 |
checkpoint kinase 1 |
3001 |
0.18 |
| chr17_42852393_42852858 | 0.46 |
ADAM11 |
ADAM metallopeptidase domain 11 |
15932 |
0.13 |
| chr14_74529404_74529555 | 0.45 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
6482 |
0.15 |
| chr6_155545820_155546032 | 0.45 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
7826 |
0.22 |
| chr3_9464516_9464980 | 0.45 |
ENSG00000266001 |
. |
3286 |
0.2 |
| chr18_46074818_46074969 | 0.45 |
CTIF |
CBP80/20-dependent translation initiation factor |
8427 |
0.26 |
| chr2_242318795_242318946 | 0.45 |
FARP2 |
FERM, RhoGEF and pleckstrin domain protein 2 |
6606 |
0.17 |
| chr8_89293881_89294032 | 0.45 |
RP11-586K2.1 |
|
45109 |
0.17 |
| chr2_174889480_174890119 | 0.45 |
SP3 |
Sp3 transcription factor |
59369 |
0.15 |
| chr2_38403806_38403957 | 0.45 |
ENSG00000199603 |
. |
29324 |
0.19 |
| chr9_124051991_124052278 | 0.45 |
GSN |
gelsolin |
3264 |
0.18 |
| chr9_124991658_124991809 | 0.45 |
LHX6 |
LIM homeobox 6 |
513 |
0.73 |
| chr5_14217746_14217971 | 0.44 |
TRIO |
trio Rho guanine nucleotide exchange factor |
33951 |
0.25 |
| chr8_37382286_37382454 | 0.44 |
RP11-150O12.6 |
|
7831 |
0.28 |
| chr10_80886709_80887280 | 0.44 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
58202 |
0.14 |
| chr7_13937610_13937761 | 0.44 |
ETV1 |
ets variant 1 |
88381 |
0.1 |
| chr1_61629231_61629382 | 0.44 |
RP4-802A10.1 |
|
38901 |
0.19 |
| chr10_4125908_4126059 | 0.44 |
KLF6 |
Kruppel-like factor 6 |
298510 |
0.01 |
| chr12_89728661_89728921 | 0.44 |
DUSP6 |
dual specificity phosphatase 6 |
16157 |
0.24 |
| chr2_197848755_197848906 | 0.44 |
ANKRD44 |
ankyrin repeat domain 44 |
16396 |
0.24 |
| chr17_40111299_40111636 | 0.44 |
CNP |
2',3'-cyclic nucleotide 3' phosphodiesterase |
7292 |
0.12 |
| chr14_59366227_59366378 | 0.43 |
ENSG00000221427 |
. |
128338 |
0.06 |
| chr6_56508196_56508347 | 0.43 |
DST |
dystonin |
477 |
0.89 |
| chr3_98575105_98575256 | 0.43 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
44835 |
0.12 |
| chr10_28976434_28976585 | 0.43 |
BAMBI |
BMP and activin membrane-bound inhibitor |
10238 |
0.19 |
| chr6_14355332_14355483 | 0.43 |
ENSG00000238987 |
. |
198714 |
0.03 |
| chr12_27265988_27266204 | 0.43 |
C12orf71 |
chromosome 12 open reading frame 71 |
30649 |
0.19 |
| chr20_56292850_56293001 | 0.43 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
6384 |
0.28 |
| chr5_136899404_136899555 | 0.43 |
ENSG00000221612 |
. |
30336 |
0.19 |
| chr2_55327280_55327923 | 0.43 |
ENSG00000266376 |
. |
8245 |
0.2 |
| chr9_112857746_112857955 | 0.42 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
5373 |
0.31 |
| chr19_47487906_47488096 | 0.42 |
NPAS1 |
neuronal PAS domain protein 1 |
35076 |
0.12 |
| chr5_71448848_71449179 | 0.42 |
ENSG00000264099 |
. |
16281 |
0.23 |
| chr3_171704686_171704902 | 0.42 |
FNDC3B |
fibronectin type III domain containing 3B |
52624 |
0.15 |
| chr16_54495686_54495837 | 0.42 |
ENSG00000264079 |
. |
87547 |
0.1 |
| chr4_22476411_22476562 | 0.42 |
GPR125 |
G protein-coupled receptor 125 |
1045 |
0.7 |
| chr12_46824110_46824422 | 0.42 |
SLC38A2 |
solute carrier family 38, member 2 |
57616 |
0.14 |
| chr10_90411662_90411938 | 0.42 |
LIPF |
lipase, gastric |
12398 |
0.19 |
| chr2_137948320_137948471 | 0.41 |
THSD7B |
thrombospondin, type I, domain containing 7B |
134121 |
0.06 |
| chr8_19963280_19963431 | 0.41 |
AC100802.3 |
|
26539 |
0.21 |
| chr1_214688078_214688229 | 0.41 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
36413 |
0.21 |
| chr13_52422853_52423004 | 0.41 |
CCDC70 |
coiled-coil domain containing 70 |
13189 |
0.2 |
| chr7_51097140_51097291 | 0.41 |
RP4-724E13.2 |
|
12055 |
0.29 |
| chr8_80260993_80261144 | 0.41 |
ENSG00000264969 |
. |
251470 |
0.02 |
| chr2_213707871_213708022 | 0.41 |
ENSG00000221388 |
. |
79547 |
0.1 |
| chr1_64434982_64435133 | 0.41 |
ENSG00000207190 |
. |
59615 |
0.14 |
| chr2_121946677_121947036 | 0.41 |
TFCP2L1 |
transcription factor CP2-like 1 |
95927 |
0.08 |
| chr1_32255908_32256059 | 0.41 |
RP11-84A19.3 |
|
1252 |
0.38 |
| chr6_134085744_134085895 | 0.41 |
RP3-323P13.2 |
|
2927 |
0.39 |
| chr3_197193061_197193212 | 0.40 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
48130 |
0.15 |
| chr6_169692535_169692779 | 0.40 |
THBS2 |
thrombospondin 2 |
38518 |
0.21 |
| chr20_21413887_21414038 | 0.40 |
ENSG00000223128 |
. |
8283 |
0.2 |
| chr15_80843805_80843956 | 0.40 |
RP11-379K22.2 |
|
11405 |
0.18 |
| chr2_19141790_19141941 | 0.40 |
NT5C1B |
5'-nucleotidase, cytosolic IB |
371027 |
0.01 |
| chr17_62258202_62258353 | 0.40 |
TEX2 |
testis expressed 2 |
31802 |
0.13 |
| chr1_214838419_214838570 | 0.40 |
CENPF |
centromere protein F, 350/400kDa |
61956 |
0.15 |
| chr2_38417733_38417884 | 0.40 |
ENSG00000199603 |
. |
43251 |
0.16 |
| chr13_81043785_81043936 | 0.40 |
ENSG00000202398 |
. |
100191 |
0.09 |
| chr6_147233902_147234394 | 0.40 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
1406 |
0.59 |
| chr10_79346896_79347047 | 0.39 |
ENSG00000199592 |
. |
164 |
0.97 |
| chr13_31293057_31293208 | 0.39 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
16513 |
0.26 |
| chr19_13162140_13162330 | 0.39 |
AC007787.2 |
|
19953 |
0.09 |
| chr14_75163942_75164093 | 0.39 |
ENSG00000222604 |
. |
14637 |
0.13 |
| chr4_84482718_84482982 | 0.39 |
AGPAT9 |
1-acylglycerol-3-phosphate O-acyltransferase 9 |
25286 |
0.2 |
| chr1_87804199_87804350 | 0.39 |
LMO4 |
LIM domain only 4 |
6923 |
0.31 |
| chr5_124392361_124392578 | 0.39 |
ENSG00000222107 |
. |
294355 |
0.01 |
| chr6_135820174_135820347 | 0.39 |
AHI1 |
Abelson helper integration site 1 |
1346 |
0.52 |
| chr18_56247923_56248074 | 0.39 |
ENSG00000252284 |
. |
19865 |
0.14 |
| chr2_182239852_182240051 | 0.39 |
ENSG00000266705 |
. |
69572 |
0.12 |
| chr14_32346804_32346955 | 0.39 |
RP11-187E13.1 |
Uncharacterized protein |
67180 |
0.12 |
| chr19_2524912_2525063 | 0.39 |
ENSG00000252962 |
. |
21848 |
0.14 |
| chr3_149096434_149096694 | 0.39 |
TM4SF1-AS1 |
TM4SF1 antisense RNA 1 |
558 |
0.59 |
| chr14_59338271_59338558 | 0.39 |
ENSG00000221427 |
. |
100450 |
0.08 |
| chr8_38896239_38896390 | 0.39 |
ENSG00000207199 |
. |
20112 |
0.16 |
| chr5_142051517_142051668 | 0.39 |
FGF1 |
fibroblast growth factor 1 (acidic) |
13645 |
0.21 |
| chr2_144711038_144711196 | 0.38 |
AC016910.1 |
|
16477 |
0.25 |
| chr11_121986232_121986383 | 0.38 |
BLID |
BH3-like motif containing, cell death inducer |
616 |
0.47 |
| chr11_27483936_27484329 | 0.38 |
RP11-159H22.2 |
|
9144 |
0.17 |
| chr6_4320523_4320712 | 0.38 |
ENSG00000201185 |
. |
107580 |
0.07 |
| chr11_123381605_123382192 | 0.38 |
GRAMD1B |
GRAM domain containing 1B |
14446 |
0.23 |
| chr5_122451826_122451977 | 0.38 |
AC106786.1 |
|
25907 |
0.15 |
| chr6_138174614_138174765 | 0.38 |
RP11-356I2.4 |
|
4496 |
0.25 |
| chr12_12377622_12377773 | 0.38 |
ENSG00000199551 |
. |
1911 |
0.38 |
| chr4_139184776_139184927 | 0.38 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
21348 |
0.27 |
| chr1_215130772_215131126 | 0.38 |
KCNK2 |
potassium channel, subfamily K, member 2 |
48249 |
0.2 |
| chr5_169372946_169373097 | 0.38 |
FAM196B |
family with sequence similarity 196, member B |
34723 |
0.2 |
| chr2_202270772_202271289 | 0.38 |
ENSG00000202008 |
. |
17206 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.3 | GO:0021853 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.1 | 0.3 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.1 | 0.2 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.2 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.3 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.2 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.2 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.6 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.6 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0031392 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0042253 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0021843 | substrate-independent telencephalic tangential migration(GO:0021826) substrate-independent telencephalic tangential interneuron migration(GO:0021843) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0071221 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.2 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0051584 | regulation of neurotransmitter uptake(GO:0051580) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.1 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.5 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.2 | GO:0030146 | obsolete diuresis(GO:0030146) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.2 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.2 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:0090659 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.2 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.0 | 0.0 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0043206 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0051532 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.0 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.1 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.1 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.2 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.1 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.0 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.0 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.0 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.0 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.1 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.0 | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process(GO:0009130) |
| 0.0 | 0.1 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) |
| 0.0 | 0.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.0 | GO:0051934 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.0 | GO:0010873 | positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 0.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 1.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.2 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.2 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.2 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.0 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |