Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for DMBX1
Z-value: 0.86
Transcription factors associated with DMBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMBX1
|
ENSG00000197587.6 | diencephalon/mesencephalon homeobox 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_46985901_46986151 | DMBX1 | 13357 | 0.144026 | 0.74 | 2.3e-02 | Click! |
| chr1_46987736_46988019 | DMBX1 | 15208 | 0.139478 | 0.74 | 2.3e-02 | Click! |
| chr1_46986402_46986650 | DMBX1 | 13857 | 0.142787 | 0.71 | 3.0e-02 | Click! |
| chr1_46985679_46985830 | DMBX1 | 13085 | 0.144705 | 0.64 | 6.4e-02 | Click! |
| chr1_46951514_46951665 | DMBX1 | 21079 | 0.150319 | -0.56 | 1.2e-01 | Click! |
Activity of the DMBX1 motif across conditions
Conditions sorted by the z-value of the DMBX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
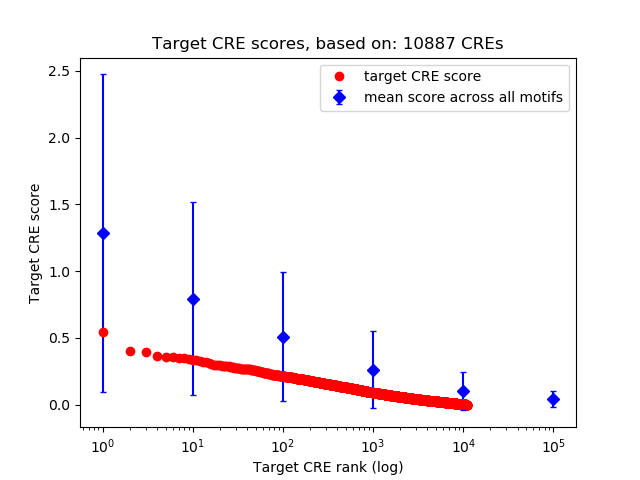
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_227289868_227290019 | 0.54 |
ENSG00000263363 |
. |
233566 |
0.02 |
| chr5_57134395_57134546 | 0.40 |
ENSG00000266864 |
. |
34403 |
0.24 |
| chr14_53301825_53302222 | 0.39 |
FERMT2 |
fermitin family member 2 |
29216 |
0.15 |
| chr12_1427689_1428011 | 0.37 |
RP5-951N9.2 |
|
67149 |
0.11 |
| chr13_45779958_45780109 | 0.36 |
KCTD4 |
potassium channel tetramerization domain containing 4 |
4858 |
0.22 |
| chr3_115504477_115504628 | 0.36 |
ENSG00000243359 |
. |
52163 |
0.18 |
| chr17_17734899_17735050 | 0.35 |
SREBF1 |
sterol regulatory element binding transcription factor 1 |
5157 |
0.15 |
| chr10_3149301_3149525 | 0.35 |
PFKP |
phosphofructokinase, platelet |
2474 |
0.34 |
| chr5_31408337_31408488 | 0.34 |
ENSG00000206682 |
. |
97379 |
0.07 |
| chr1_201426596_201426747 | 0.34 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
11641 |
0.15 |
| chr5_38871423_38871574 | 0.34 |
OSMR |
oncostatin M receptor |
25397 |
0.24 |
| chr18_46148945_46149096 | 0.32 |
ENSG00000266276 |
. |
47951 |
0.15 |
| chr8_21614873_21615024 | 0.32 |
GFRA2 |
GDNF family receptor alpha 2 |
30887 |
0.22 |
| chr4_182470842_182470993 | 0.32 |
ENSG00000251742 |
. |
285344 |
0.01 |
| chr1_18201641_18201961 | 0.31 |
ACTL8 |
actin-like 8 |
119993 |
0.06 |
| chr2_224966339_224966634 | 0.30 |
SERPINE2 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
62450 |
0.13 |
| chr9_83699073_83699224 | 0.30 |
ENSG00000221581 |
. |
349904 |
0.01 |
| chr2_87553379_87553697 | 0.30 |
IGKV3OR2-268 |
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
12096 |
0.29 |
| chr16_31076495_31076764 | 0.30 |
AC135050.5 |
|
187 |
0.51 |
| chr11_73849362_73849513 | 0.29 |
C2CD3 |
C2 calcium-dependent domain containing 3 |
32592 |
0.14 |
| chr21_44937001_44937451 | 0.29 |
SIK1 |
salt-inducible kinase 1 |
90218 |
0.08 |
| chr8_13340149_13340300 | 0.29 |
DLC1 |
deleted in liver cancer 1 |
16526 |
0.23 |
| chr1_113088459_113088610 | 0.29 |
RP4-671G15.2 |
|
27471 |
0.14 |
| chr10_54078584_54078735 | 0.29 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
4603 |
0.27 |
| chr16_50219117_50219379 | 0.29 |
PAPD5 |
PAP associated domain containing 5 |
31469 |
0.14 |
| chr4_122591083_122591234 | 0.28 |
ANXA5 |
annexin A5 |
26959 |
0.21 |
| chr11_19956797_19957009 | 0.28 |
NAV2-AS3 |
NAV2 antisense RNA 3 |
45980 |
0.17 |
| chr10_35021138_35021390 | 0.28 |
PARD3 |
par-3 family cell polarity regulator |
82985 |
0.1 |
| chr17_17840585_17840808 | 0.28 |
TOM1L2 |
target of myb1-like 2 (chicken) |
35015 |
0.12 |
| chr10_79291019_79291170 | 0.28 |
ENSG00000199592 |
. |
55713 |
0.15 |
| chr15_60671550_60671832 | 0.27 |
ANXA2 |
annexin A2 |
4987 |
0.3 |
| chr6_152412861_152413012 | 0.27 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
76550 |
0.12 |
| chr9_14278008_14278159 | 0.27 |
NFIB |
nuclear factor I/B |
29929 |
0.22 |
| chr17_53803344_53803495 | 0.27 |
TMEM100 |
transmembrane protein 100 |
3202 |
0.32 |
| chr3_29736769_29736920 | 0.27 |
RBMS3-AS2 |
RBMS3 antisense RNA 2 |
52544 |
0.18 |
| chr12_49607723_49607874 | 0.27 |
TUBA1C |
tubulin, alpha 1c |
13911 |
0.12 |
| chr12_80987199_80987385 | 0.27 |
RP11-272K23.3 |
|
188 |
0.96 |
| chr1_8078544_8078695 | 0.27 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
2926 |
0.27 |
| chr16_71963323_71963474 | 0.27 |
RP11-498D10.6 |
|
516 |
0.69 |
| chr8_119017458_119017609 | 0.27 |
EXT1 |
exostosin glycosyltransferase 1 |
105120 |
0.08 |
| chr19_47478570_47478952 | 0.26 |
ENSG00000252071 |
. |
27215 |
0.14 |
| chr10_75277605_75277756 | 0.26 |
USP54 |
ubiquitin specific peptidase 54 |
5752 |
0.13 |
| chr3_188066250_188066414 | 0.26 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
108680 |
0.07 |
| chr2_112289666_112289908 | 0.26 |
ENSG00000266139 |
. |
211119 |
0.02 |
| chr5_159969265_159969416 | 0.26 |
ENSG00000253522 |
. |
56981 |
0.1 |
| chr3_119864314_119864465 | 0.26 |
ENSG00000244139 |
. |
23633 |
0.17 |
| chr9_20319925_20320191 | 0.26 |
ENSG00000221744 |
. |
25056 |
0.21 |
| chr6_138409190_138409341 | 0.26 |
PERP |
PERP, TP53 apoptosis effector |
19383 |
0.24 |
| chr8_117698330_117698481 | 0.26 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
69618 |
0.11 |
| chr2_109968074_109968225 | 0.26 |
ENSG00000265965 |
. |
38068 |
0.22 |
| chr1_182367363_182367514 | 0.25 |
TEDDM1 |
transmembrane epididymal protein 1 |
2313 |
0.3 |
| chr10_106094941_106095155 | 0.25 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
1385 |
0.33 |
| chr7_46972074_46972225 | 0.25 |
AC011294.3 |
Uncharacterized protein |
235429 |
0.02 |
| chr14_69544397_69544681 | 0.25 |
ENSG00000206768 |
. |
66131 |
0.1 |
| chr19_47756899_47757050 | 0.25 |
CCDC9 |
coiled-coil domain containing 9 |
2742 |
0.19 |
| chr15_41990797_41991061 | 0.25 |
ENSG00000207766 |
. |
7146 |
0.17 |
| chr9_34648155_34648409 | 0.25 |
GALT |
galactose-1-phosphate uridylyltransferase |
1622 |
0.17 |
| chr3_149336105_149336256 | 0.25 |
WWTR1-IT1 |
WWTR1 intronic transcript 1 (non-protein coding) |
31791 |
0.14 |
| chr5_171708669_171708924 | 0.25 |
UBTD2 |
ubiquitin domain containing 2 |
2279 |
0.25 |
| chr17_77032976_77033127 | 0.24 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
2784 |
0.2 |
| chr13_53033095_53033246 | 0.24 |
CKAP2 |
cytoskeleton associated protein 2 |
3027 |
0.25 |
| chr16_14597322_14597496 | 0.24 |
AC092291.2 |
|
32567 |
0.18 |
| chr10_33418933_33419614 | 0.24 |
ENSG00000263576 |
. |
31709 |
0.18 |
| chr13_44842332_44842483 | 0.24 |
SERP2 |
stress-associated endoplasmic reticulum protein family member 2 |
105571 |
0.06 |
| chr15_36761585_36761736 | 0.24 |
C15orf41 |
chromosome 15 open reading frame 41 |
110152 |
0.08 |
| chr2_192529911_192530062 | 0.24 |
NABP1 |
nucleic acid binding protein 1 |
12876 |
0.29 |
| chr5_156943457_156943608 | 0.24 |
ADAM19 |
ADAM metallopeptidase domain 19 |
13702 |
0.17 |
| chr12_66204140_66204291 | 0.24 |
HMGA2 |
high mobility group AT-hook 2 |
13696 |
0.22 |
| chr3_48099925_48100076 | 0.23 |
MAP4 |
microtubule-associated protein 4 |
30314 |
0.15 |
| chr1_193155079_193155230 | 0.23 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
630 |
0.75 |
| chr18_47812755_47812906 | 0.23 |
CXXC1 |
CXXC finger protein 1 |
1110 |
0.44 |
| chr17_79855827_79855978 | 0.23 |
NPB |
neuropeptide B |
2719 |
0.08 |
| chr9_4243033_4243184 | 0.23 |
GLIS3 |
GLIS family zinc finger 3 |
55388 |
0.13 |
| chr10_73292337_73292498 | 0.23 |
CDH23-AS1 |
CDH23 antisense RNA 1 |
20787 |
0.22 |
| chr8_23411869_23412065 | 0.23 |
AC051642.1 |
|
8684 |
0.16 |
| chr12_50640493_50640685 | 0.23 |
LIMA1 |
LIM domain and actin binding 1 |
3087 |
0.17 |
| chr13_52946332_52946483 | 0.23 |
ENSG00000199605 |
. |
27732 |
0.16 |
| chr5_39014905_39015056 | 0.23 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
59511 |
0.14 |
| chr17_38612271_38612422 | 0.22 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
12633 |
0.13 |
| chr1_198243273_198243424 | 0.22 |
NEK7 |
NIMA-related kinase 7 |
53419 |
0.18 |
| chr18_20693944_20694095 | 0.22 |
CABLES1 |
Cdk5 and Abl enzyme substrate 1 |
20509 |
0.18 |
| chr2_226981146_226981297 | 0.22 |
ENSG00000263363 |
. |
542288 |
0.0 |
| chr9_95269405_95269571 | 0.22 |
ENSG00000264158 |
. |
20852 |
0.15 |
| chr13_110665079_110665230 | 0.22 |
ENSG00000265885 |
. |
85344 |
0.1 |
| chr11_35693192_35693343 | 0.22 |
TRIM44 |
tripartite motif containing 44 |
8914 |
0.25 |
| chr12_90485340_90485587 | 0.22 |
ENSG00000252823 |
. |
337627 |
0.01 |
| chr1_211210156_211210307 | 0.22 |
KCNH1-IT1 |
KCNH1 intronic transcript 1 (non-protein coding) |
96485 |
0.08 |
| chr11_8855031_8855440 | 0.22 |
ST5 |
suppression of tumorigenicity 5 |
1749 |
0.28 |
| chr7_28058858_28059009 | 0.22 |
JAZF1 |
JAZF zinc finger 1 |
1485 |
0.53 |
| chr10_3570543_3570694 | 0.22 |
RP11-184A2.3 |
|
222641 |
0.02 |
| chr9_13750648_13750799 | 0.22 |
NFIB |
nuclear factor I/B |
430068 |
0.01 |
| chr5_112606912_112607063 | 0.22 |
MCC |
mutated in colorectal cancers |
23659 |
0.23 |
| chr9_683590_683741 | 0.22 |
RP11-130C19.3 |
|
1890 |
0.39 |
| chr12_56734029_56734180 | 0.22 |
IL23A |
interleukin 23, alpha subunit p19 |
1441 |
0.19 |
| chr4_169142842_169142993 | 0.22 |
DDX60 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
14496 |
0.26 |
| chr11_95609659_95609810 | 0.22 |
ENSG00000222578 |
. |
36446 |
0.16 |
| chr7_46987783_46987934 | 0.22 |
AC011294.3 |
Uncharacterized protein |
251138 |
0.02 |
| chr6_81251109_81251260 | 0.22 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
434801 |
0.01 |
| chrX_17632522_17632673 | 0.22 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
20816 |
0.22 |
| chr5_106717368_106717590 | 0.21 |
EFNA5 |
ephrin-A5 |
288849 |
0.01 |
| chr1_85805501_85805652 | 0.21 |
ENSG00000264380 |
. |
55283 |
0.09 |
| chr16_30887666_30887829 | 0.21 |
MIR4519 |
microRNA 4519 |
514 |
0.52 |
| chr5_168650940_168651091 | 0.21 |
ENSG00000207619 |
. |
39683 |
0.19 |
| chr8_42949619_42950327 | 0.21 |
POMK |
protein-O-mannose kinase |
1312 |
0.4 |
| chr3_189721675_189721826 | 0.21 |
ENSG00000265045 |
. |
109973 |
0.06 |
| chr8_124562512_124562663 | 0.21 |
FBXO32 |
F-box protein 32 |
9141 |
0.2 |
| chr16_66652366_66652517 | 0.21 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
13017 |
0.11 |
| chr2_219576440_219576655 | 0.21 |
TTLL4 |
tubulin tyrosine ligase-like family, member 4 |
830 |
0.51 |
| chr17_39524192_39524343 | 0.21 |
KRT33B |
keratin 33B |
1785 |
0.18 |
| chr8_118860902_118861053 | 0.21 |
EXT1 |
exostosin glycosyltransferase 1 |
261676 |
0.02 |
| chr9_81627686_81627966 | 0.21 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
558862 |
0.0 |
| chr6_170715097_170715248 | 0.21 |
RP1-140C12.2 |
|
8055 |
0.25 |
| chr4_95749215_95749366 | 0.21 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
70171 |
0.14 |
| chr18_26091967_26092118 | 0.21 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
334632 |
0.01 |
| chr3_29555190_29555652 | 0.21 |
RBMS3-AS2 |
RBMS3 antisense RNA 2 |
103125 |
0.08 |
| chr1_61563332_61563483 | 0.21 |
NFIA |
nuclear factor I/A |
9323 |
0.21 |
| chr6_138153686_138153837 | 0.21 |
RP11-356I2.4 |
|
6730 |
0.23 |
| chr7_17599104_17599255 | 0.21 |
ENSG00000199473 |
. |
186229 |
0.03 |
| chr3_27583890_27584041 | 0.21 |
ENSG00000238912 |
. |
5384 |
0.24 |
| chr8_54797197_54797453 | 0.21 |
RGS20 |
regulator of G-protein signaling 20 |
3871 |
0.22 |
| chr17_57435169_57435320 | 0.21 |
ENSG00000263857 |
. |
8200 |
0.17 |
| chr6_146680396_146680547 | 0.21 |
ENSG00000222971 |
. |
18915 |
0.29 |
| chr2_187467884_187468035 | 0.21 |
ITGAV |
integrin, alpha V |
3027 |
0.32 |
| chr1_68851372_68851523 | 0.21 |
RPE65 |
retinal pigment epithelium-specific protein 65kDa |
64195 |
0.12 |
| chr3_193696417_193696568 | 0.20 |
RP11-135A1.2 |
|
152498 |
0.04 |
| chr18_55439591_55439742 | 0.20 |
ENSG00000202159 |
. |
17040 |
0.19 |
| chr2_192759273_192759424 | 0.20 |
AC098617.1 |
|
27924 |
0.21 |
| chr18_42748060_42748211 | 0.20 |
RP11-846C15.2 |
|
12944 |
0.26 |
| chr11_11048560_11048711 | 0.20 |
ZBED5-AS1 |
ZBED5 antisense RNA 1 |
161827 |
0.03 |
| chr3_89035703_89035854 | 0.20 |
EPHA3 |
EPH receptor A3 |
120896 |
0.07 |
| chr1_26344271_26344422 | 0.20 |
EXTL1 |
exostosin-like glycosyltransferase 1 |
3925 |
0.12 |
| chr18_56345406_56345557 | 0.20 |
RP11-126O1.4 |
|
635 |
0.66 |
| chr10_120710049_120710200 | 0.20 |
RP11-498J9.2 |
|
53272 |
0.11 |
| chr3_189555786_189555937 | 0.20 |
ENSG00000216058 |
. |
8150 |
0.28 |
| chr3_16490582_16490733 | 0.20 |
RFTN1 |
raftlin, lipid raft linker 1 |
33715 |
0.19 |
| chr17_5326160_5326311 | 0.20 |
RPAIN |
RPA interacting protein |
2713 |
0.15 |
| chr7_22711550_22711701 | 0.20 |
AC002480.5 |
|
10214 |
0.19 |
| chr6_20876608_20876759 | 0.20 |
RP3-348I23.2 |
|
75758 |
0.12 |
| chr3_45177863_45178014 | 0.20 |
CDCP1 |
CUB domain containing protein 1 |
9976 |
0.23 |
| chr11_35181391_35181660 | 0.20 |
CD44 |
CD44 molecule (Indian blood group) |
16593 |
0.15 |
| chr16_82688055_82688226 | 0.20 |
CDH13 |
cadherin 13 |
27442 |
0.24 |
| chr1_219497938_219498089 | 0.20 |
RP11-135J2.4 |
|
150710 |
0.04 |
| chr5_171384729_171385179 | 0.20 |
FBXW11 |
F-box and WD repeat domain containing 11 |
19806 |
0.24 |
| chr1_146638682_146638833 | 0.19 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
5289 |
0.17 |
| chr10_22888353_22888956 | 0.19 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
8012 |
0.31 |
| chr12_64855072_64855223 | 0.19 |
TBK1 |
TANK-binding kinase 1 |
8892 |
0.2 |
| chr4_151049516_151049667 | 0.19 |
DCLK2 |
doublecortin-like kinase 2 |
49411 |
0.17 |
| chr15_99881623_99881774 | 0.19 |
AC022819.2 |
Uncharacterized protein |
21870 |
0.18 |
| chr15_71087954_71088105 | 0.19 |
RP11-138H8.3 |
|
16822 |
0.17 |
| chr5_123287126_123287277 | 0.19 |
CSNK1G3 |
casein kinase 1, gamma 3 |
363082 |
0.01 |
| chr10_63803048_63803479 | 0.19 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
5707 |
0.3 |
| chr7_116417479_116417780 | 0.19 |
MET |
met proto-oncogene |
400 |
0.88 |
| chr1_185614085_185614236 | 0.19 |
ENSG00000201596 |
. |
10846 |
0.23 |
| chr1_150481830_150481981 | 0.19 |
ECM1 |
extracellular matrix protein 1 |
1322 |
0.29 |
| chr17_1283034_1283185 | 0.19 |
YWHAE |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon |
18521 |
0.16 |
| chr12_118628458_118628609 | 0.19 |
TAOK3 |
TAO kinase 3 |
177 |
0.96 |
| chr12_49315908_49316059 | 0.19 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
2767 |
0.11 |
| chr1_87243934_87244085 | 0.19 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
73431 |
0.09 |
| chr12_52622387_52622538 | 0.19 |
KRT7 |
keratin 7 |
4436 |
0.12 |
| chr5_73926315_73926659 | 0.19 |
HEXB |
hexosaminidase B (beta polypeptide) |
9361 |
0.21 |
| chr8_119392429_119392580 | 0.19 |
AC023590.1 |
Uncharacterized protein |
98023 |
0.09 |
| chr12_46836665_46836816 | 0.19 |
SLC38A2 |
solute carrier family 38, member 2 |
70090 |
0.12 |
| chr15_79973577_79973728 | 0.19 |
ENSG00000252995 |
. |
136140 |
0.05 |
| chr2_65619842_65619993 | 0.19 |
AC012370.2 |
|
12098 |
0.22 |
| chr12_56119220_56119371 | 0.19 |
CD63 |
CD63 molecule |
1548 |
0.18 |
| chr12_52182216_52182367 | 0.19 |
AC068987.1 |
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
21498 |
0.2 |
| chr17_18626006_18626157 | 0.19 |
TRIM16L |
tripartite motif containing 16-like |
647 |
0.64 |
| chr5_94828359_94828510 | 0.19 |
ENSG00000212259 |
. |
28639 |
0.17 |
| chr3_40085086_40085237 | 0.19 |
MYRIP |
myosin VIIA and Rab interacting protein |
56341 |
0.14 |
| chr13_109926641_109926792 | 0.19 |
MYO16-AS1 |
MYO16 antisense RNA 1 |
72885 |
0.13 |
| chr13_40106261_40106431 | 0.19 |
LHFP |
lipoma HMGIC fusion partner |
70962 |
0.11 |
| chr9_91362565_91362744 | 0.19 |
ENSG00000265873 |
. |
1834 |
0.5 |
| chr10_21695519_21695777 | 0.19 |
ENSG00000199222 |
. |
74997 |
0.08 |
| chr6_79520805_79520956 | 0.19 |
IRAK1BP1 |
interleukin-1 receptor-associated kinase 1 binding protein 1 |
56309 |
0.17 |
| chr2_102420275_102420522 | 0.19 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
6672 |
0.3 |
| chr9_116103856_116104007 | 0.19 |
WDR31 |
WD repeat domain 31 |
1353 |
0.33 |
| chr16_30079690_30079841 | 0.19 |
ALDOA |
aldolase A, fructose-bisphosphate |
941 |
0.31 |
| chr6_155815833_155816170 | 0.19 |
NOX3 |
NADPH oxidase 3 |
38964 |
0.21 |
| chr11_111429197_111429348 | 0.19 |
LAYN |
layilin |
16995 |
0.12 |
| chr2_3626762_3626913 | 0.19 |
ENSG00000252531 |
. |
1323 |
0.29 |
| chr11_10372140_10372291 | 0.19 |
AMPD3 |
adenosine monophosphate deaminase 3 |
32997 |
0.13 |
| chr9_74367560_74367715 | 0.19 |
TMEM2 |
transmembrane protein 2 |
15665 |
0.28 |
| chr12_27233938_27234089 | 0.19 |
C12orf71 |
chromosome 12 open reading frame 71 |
1434 |
0.46 |
| chr3_129460063_129460214 | 0.19 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
52563 |
0.12 |
| chr4_17582411_17582562 | 0.18 |
LAP3 |
leucine aminopeptidase 3 |
2739 |
0.22 |
| chr11_100663032_100663183 | 0.18 |
ENSG00000265208 |
. |
2065 |
0.39 |
| chr17_15859629_15859837 | 0.18 |
ADORA2B |
adenosine A2b receptor |
11502 |
0.17 |
| chr15_33136245_33136396 | 0.18 |
FMN1 |
formin 1 |
44135 |
0.15 |
| chr12_13254871_13255022 | 0.18 |
GSG1 |
germ cell associated 1 |
1630 |
0.41 |
| chr1_84355429_84355580 | 0.18 |
TTLL7-IT1 |
TTLL7 intronic transcript 1 (non-protein coding) |
94479 |
0.08 |
| chr11_10929627_10929778 | 0.18 |
ZBED5-AS1 |
ZBED5 antisense RNA 1 |
42894 |
0.14 |
| chr3_123979690_123979841 | 0.18 |
KALRN |
kalirin, RhoGEF kinase |
32928 |
0.22 |
| chr6_39855140_39855437 | 0.18 |
RP11-61I13.3 |
|
944 |
0.63 |
| chr6_132520639_132520790 | 0.18 |
ENSG00000265669 |
. |
84311 |
0.1 |
| chr11_12011920_12012071 | 0.18 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
18135 |
0.26 |
| chr10_111651373_111651848 | 0.18 |
XPNPEP1 |
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
15967 |
0.2 |
| chr11_58161507_58161658 | 0.18 |
OR5B3 |
olfactory receptor, family 5, subfamily B, member 3 |
9300 |
0.17 |
| chr7_105857899_105858050 | 0.18 |
ENSG00000201796 |
. |
9361 |
0.25 |
| chr1_40732198_40732349 | 0.18 |
ZMPSTE24 |
zinc metallopeptidase STE24 |
8494 |
0.14 |
| chr2_40175326_40175477 | 0.18 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
28568 |
0.26 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.2 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.2 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0032909 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.2 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0061043 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0070168 | negative regulation of biomineral tissue development(GO:0070168) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.0 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.0 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.2 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.0 | GO:0017000 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0033483 | gas homeostasis(GO:0033483) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.0 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.0 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.0 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0075713 | establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.0 | GO:0032347 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.0 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.2 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.0 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.0 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |