Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
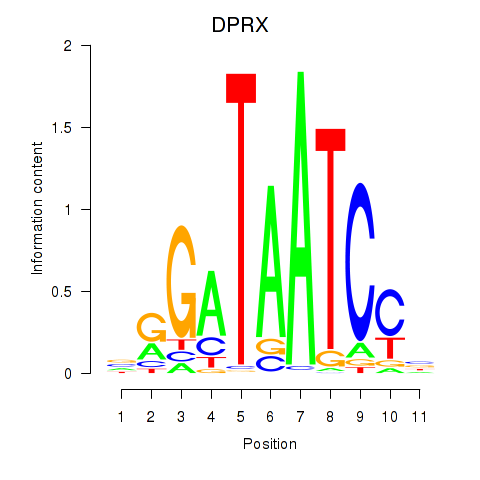
Results for DPRX
Z-value: 1.92
Transcription factors associated with DPRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DPRX
|
ENSG00000204595.1 | divergent-paired related homeobox |
Activity of the DPRX motif across conditions
Conditions sorted by the z-value of the DPRX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
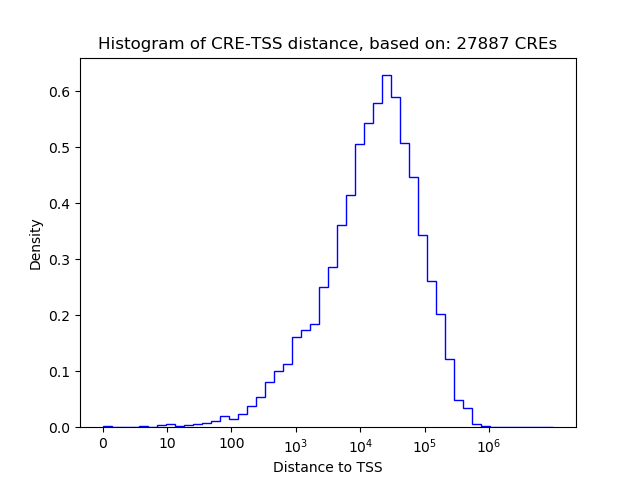
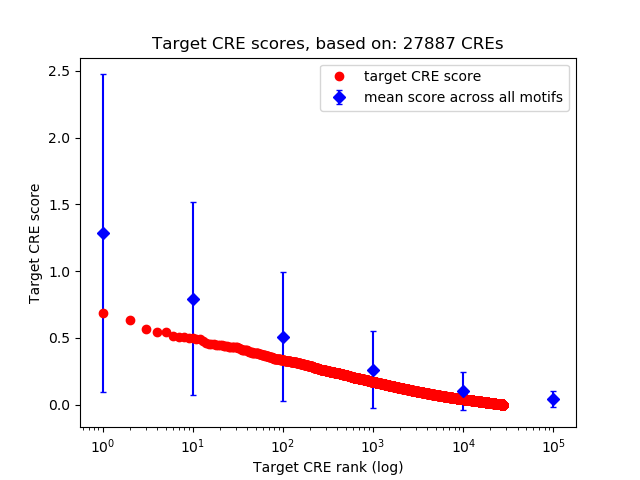
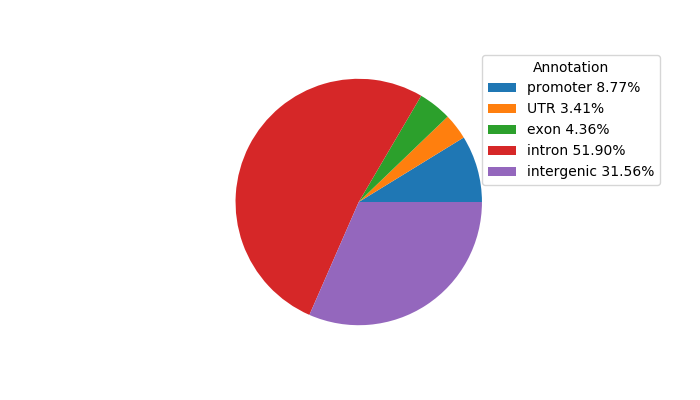
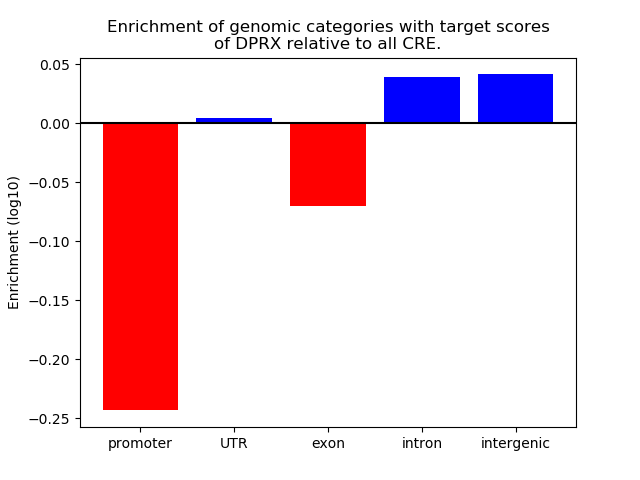
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_121906222_121906519 | 0.68 |
ENSG00000252556 |
. |
31307 |
0.16 |
| chr17_47900739_47901025 | 0.63 |
RP11-304F15.3 |
|
22390 |
0.13 |
| chr12_4140465_4141124 | 0.56 |
RP11-664D1.1 |
|
126408 |
0.05 |
| chr1_100789092_100789243 | 0.54 |
CDC14A |
cell division cycle 14A |
21417 |
0.15 |
| chr5_102310797_102310948 | 0.54 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
25591 |
0.26 |
| chr2_145266955_145267602 | 0.52 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
7837 |
0.25 |
| chr1_56185258_56185409 | 0.50 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
219620 |
0.02 |
| chr7_107331221_107331712 | 0.50 |
SLC26A4 |
solute carrier family 26 (anion exchanger), member 4 |
869 |
0.57 |
| chr15_101807298_101807582 | 0.50 |
VIMP |
VCP-interacting membrane protein |
10052 |
0.15 |
| chr10_79267255_79267533 | 0.50 |
ENSG00000199592 |
. |
79413 |
0.1 |
| chr19_10529958_10530965 | 0.49 |
CDC37 |
cell division cycle 37 |
336 |
0.7 |
| chr16_640639_641593 | 0.49 |
RAB40C |
RAB40C, member RAS oncogene family |
784 |
0.37 |
| chr22_44759775_44760116 | 0.48 |
RP1-32I10.10 |
Uncharacterized protein |
1486 |
0.51 |
| chr15_43806030_43806788 | 0.46 |
MAP1A |
microtubule-associated protein 1A |
3253 |
0.18 |
| chr3_193988294_193988600 | 0.46 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
83600 |
0.08 |
| chr12_94312317_94312468 | 0.46 |
RP11-1060G2.1 |
|
24987 |
0.2 |
| chr1_110923779_110923930 | 0.45 |
LAMTOR5-AS1 |
LAMTOR5 antisense RNA 1 |
1587 |
0.24 |
| chr16_66150653_66150937 | 0.45 |
ENSG00000201999 |
. |
184917 |
0.03 |
| chr8_25771009_25771433 | 0.45 |
EBF2 |
early B-cell factor 2 |
25789 |
0.27 |
| chr6_45619110_45619261 | 0.45 |
ENSG00000252738 |
. |
5344 |
0.34 |
| chr14_69015321_69015580 | 0.45 |
CTD-2325P2.4 |
|
79712 |
0.1 |
| chr19_41035362_41036035 | 0.44 |
SPTBN4 |
spectrin, beta, non-erythrocytic 4 |
697 |
0.62 |
| chr1_219761600_219761751 | 0.44 |
ENSG00000252240 |
. |
75044 |
0.12 |
| chr9_111202576_111202727 | 0.44 |
ENSG00000222512 |
. |
81442 |
0.11 |
| chr4_148693217_148693552 | 0.43 |
ENSG00000264274 |
. |
10362 |
0.18 |
| chr5_143022321_143022472 | 0.43 |
ENSG00000266478 |
. |
37029 |
0.18 |
| chr11_131626567_131626718 | 0.43 |
NTM |
neurotrimin |
95754 |
0.08 |
| chr1_146736722_146736873 | 0.43 |
CHD1L |
chromodomain helicase DNA binding protein 1-like |
22441 |
0.19 |
| chr21_36625750_36625901 | 0.43 |
RUNX1 |
runt-related transcription factor 1 |
204184 |
0.03 |
| chr17_57502582_57502794 | 0.43 |
RP11-567L7.5 |
|
22024 |
0.18 |
| chr5_118728587_118729031 | 0.43 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
37079 |
0.14 |
| chr6_143169141_143169292 | 0.42 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
11032 |
0.28 |
| chr12_110388287_110388838 | 0.42 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
45467 |
0.11 |
| chr1_203147074_203147225 | 0.42 |
MYBPH |
myosin binding protein H |
2208 |
0.22 |
| chr1_120485813_120485964 | 0.41 |
RP5-1042I8.7 |
|
32652 |
0.18 |
| chr9_94579747_94580327 | 0.41 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
131125 |
0.05 |
| chr3_197238562_197238713 | 0.41 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
2629 |
0.37 |
| chr10_44973931_44974082 | 0.41 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
93466 |
0.08 |
| chr16_87120298_87120449 | 0.41 |
RP11-899L11.3 |
|
129148 |
0.05 |
| chr1_153521085_153521570 | 0.41 |
S100A3 |
S100 calcium binding protein A3 |
275 |
0.76 |
| chr3_159740833_159741097 | 0.40 |
LINC01100 |
long intergenic non-protein coding RNA 1100 |
7154 |
0.2 |
| chr4_72061466_72061617 | 0.40 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
8497 |
0.31 |
| chr17_41487227_41487378 | 0.40 |
ENSG00000251763 |
. |
2493 |
0.17 |
| chr2_46196026_46196177 | 0.39 |
PRKCE |
protein kinase C, epsilon |
31940 |
0.23 |
| chr7_132423668_132423819 | 0.39 |
ENSG00000201009 |
. |
14040 |
0.21 |
| chr3_123836876_123837056 | 0.39 |
ENSG00000266383 |
. |
14810 |
0.2 |
| chr12_76636613_76636764 | 0.39 |
ENSG00000223273 |
. |
72843 |
0.1 |
| chr8_98910268_98910419 | 0.39 |
MATN2 |
matrilin 2 |
10138 |
0.24 |
| chr1_120508182_120508333 | 0.39 |
RP5-1042I8.7 |
|
55021 |
0.13 |
| chr9_88955484_88955635 | 0.39 |
ZCCHC6 |
zinc finger, CCHC domain containing 6 |
2479 |
0.34 |
| chr4_77657284_77657435 | 0.39 |
RP11-359D14.2 |
|
22268 |
0.2 |
| chr2_191910102_191910403 | 0.39 |
ENSG00000231858 |
. |
24000 |
0.15 |
| chr17_59383316_59383467 | 0.38 |
RP11-332H18.3 |
|
76755 |
0.08 |
| chr3_100730226_100730377 | 0.38 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
17942 |
0.25 |
| chr11_75338227_75338378 | 0.38 |
CTD-2530H12.7 |
|
11330 |
0.14 |
| chr6_131221994_131222145 | 0.38 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
10505 |
0.29 |
| chr2_63991472_63991650 | 0.38 |
ENSG00000221085 |
. |
68892 |
0.11 |
| chr5_158232307_158232458 | 0.38 |
CTD-2363C16.1 |
|
177632 |
0.03 |
| chr3_16225459_16225610 | 0.37 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
9318 |
0.25 |
| chr1_219512383_219512534 | 0.37 |
RP11-135J2.4 |
|
165155 |
0.04 |
| chr7_111079181_111079332 | 0.37 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
46285 |
0.2 |
| chr21_28335085_28335236 | 0.37 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
3672 |
0.27 |
| chr13_21653666_21653823 | 0.37 |
LATS2 |
large tumor suppressor kinase 2 |
18058 |
0.14 |
| chr2_237654687_237654838 | 0.37 |
ACKR3 |
atypical chemokine receptor 3 |
176478 |
0.03 |
| chr2_37797708_37797859 | 0.37 |
AC006369.2 |
|
29496 |
0.21 |
| chr11_130717130_130717402 | 0.36 |
SNX19 |
sorting nexin 19 |
46503 |
0.19 |
| chrX_19655066_19655217 | 0.36 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
33339 |
0.23 |
| chr8_108344894_108345045 | 0.36 |
ANGPT1 |
angiopoietin 1 |
3781 |
0.36 |
| chr11_26798911_26799062 | 0.36 |
SLC5A12 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
54078 |
0.17 |
| chr14_58313511_58313662 | 0.36 |
ENSG00000252837 |
. |
9208 |
0.24 |
| chr3_79211716_79211867 | 0.36 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
143182 |
0.05 |
| chr3_120150563_120150714 | 0.36 |
FSTL1 |
follistatin-like 1 |
19200 |
0.23 |
| chr2_55346845_55347305 | 0.36 |
RTN4 |
reticulon 4 |
7318 |
0.2 |
| chr12_43684777_43685026 | 0.36 |
ENSG00000215993 |
. |
482 |
0.89 |
| chr5_39474112_39474263 | 0.36 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
11785 |
0.3 |
| chr11_101312154_101312305 | 0.35 |
ENSG00000263885 |
. |
78407 |
0.11 |
| chr17_61434785_61434936 | 0.35 |
AC037445.1 |
|
18446 |
0.15 |
| chr16_70748567_70748858 | 0.35 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
14564 |
0.14 |
| chr3_61557704_61557855 | 0.35 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
10194 |
0.32 |
| chr14_63983384_63983535 | 0.35 |
PPP2R5E |
protein phosphatase 2, regulatory subunit B', epsilon isoform |
8504 |
0.16 |
| chr2_200235271_200235422 | 0.35 |
RP11-486F17.1 |
|
45313 |
0.17 |
| chr11_78029619_78030164 | 0.35 |
RP11-452H21.1 |
|
5913 |
0.21 |
| chr15_75757189_75757340 | 0.34 |
SIN3A |
SIN3 transcription regulator family member A |
9081 |
0.18 |
| chr5_169029408_169029559 | 0.34 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
11375 |
0.22 |
| chr8_53030633_53030784 | 0.34 |
RP11-26M5.3 |
|
32672 |
0.21 |
| chr3_179519553_179519750 | 0.34 |
PEX5L-AS1 |
PEX5L antisense RNA 1 |
73513 |
0.09 |
| chr1_178358349_178358628 | 0.34 |
RASAL2 |
RAS protein activator like 2 |
47882 |
0.16 |
| chr7_25891719_25892347 | 0.34 |
ENSG00000199085 |
. |
97573 |
0.09 |
| chr12_15843418_15843825 | 0.34 |
ENSG00000200105 |
. |
18877 |
0.22 |
| chr18_18585315_18585510 | 0.34 |
ROCK1 |
Rho-associated, coiled-coil containing protein kinase 1 |
50581 |
0.15 |
| chr3_73510661_73510812 | 0.34 |
PDZRN3 |
PDZ domain containing ring finger 3 |
13047 |
0.29 |
| chr12_22066370_22066521 | 0.34 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
2533 |
0.37 |
| chr2_189540912_189541063 | 0.34 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
106177 |
0.07 |
| chr7_48222386_48222537 | 0.34 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
11404 |
0.28 |
| chr15_71335826_71335977 | 0.34 |
THSD4 |
thrombospondin, type I, domain containing 4 |
53390 |
0.14 |
| chr10_129721465_129721616 | 0.34 |
RP11-4C20.4 |
|
11422 |
0.21 |
| chr1_86953450_86953601 | 0.34 |
CLCA1 |
chloride channel accessory 1 |
18999 |
0.2 |
| chr3_139028342_139028493 | 0.34 |
MRPS22 |
mitochondrial ribosomal protein S22 |
34444 |
0.14 |
| chr8_98001138_98001426 | 0.34 |
CPQ |
carboxypeptidase Q |
40415 |
0.21 |
| chr19_1361317_1362052 | 0.33 |
MUM1 |
melanoma associated antigen (mutated) 1 |
5321 |
0.1 |
| chr6_113468632_113468783 | 0.33 |
ENSG00000201386 |
. |
176012 |
0.03 |
| chr11_128685529_128685949 | 0.33 |
KCNJ1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
26690 |
0.17 |
| chr2_68351761_68351912 | 0.33 |
WDR92 |
WD repeat domain 92 |
14081 |
0.18 |
| chr7_20204502_20204823 | 0.33 |
MACC1 |
metastasis associated in colon cancer 1 |
20039 |
0.18 |
| chr3_38870449_38870600 | 0.33 |
RP11-134J21.1 |
|
5131 |
0.21 |
| chr8_72778454_72778605 | 0.33 |
MSC |
musculin |
21826 |
0.17 |
| chr8_9305826_9305977 | 0.33 |
ENSG00000201815 |
. |
77196 |
0.1 |
| chr11_72868697_72869453 | 0.33 |
FCHSD2 |
FCH and double SH3 domains 2 |
15769 |
0.19 |
| chr10_96075798_96075949 | 0.33 |
RP11-76P2.4 |
|
789 |
0.65 |
| chr5_119879644_119879795 | 0.33 |
PRR16 |
proline rich 16 |
12560 |
0.31 |
| chr14_75295916_75296067 | 0.33 |
YLPM1 |
YLP motif containing 1 |
12050 |
0.14 |
| chr6_52406315_52406466 | 0.33 |
TRAM2 |
translocation associated membrane protein 2 |
35323 |
0.19 |
| chr10_114819664_114819822 | 0.33 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
83936 |
0.1 |
| chr7_27498955_27499106 | 0.33 |
HIBADH |
3-hydroxyisobutyrate dehydrogenase |
188345 |
0.02 |
| chr11_73578174_73578325 | 0.33 |
COA4 |
cytochrome c oxidase assembly factor 4 homolog (S. cerevisiae) |
7271 |
0.15 |
| chr14_75085505_75086322 | 0.33 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
6607 |
0.2 |
| chr2_121583330_121583481 | 0.33 |
GLI2 |
GLI family zinc finger 2 |
28484 |
0.25 |
| chr7_44923182_44923985 | 0.33 |
RP4-673M15.1 |
|
969 |
0.35 |
| chr5_169289355_169289506 | 0.32 |
CTB-37A13.1 |
|
83061 |
0.09 |
| chr5_151052659_151052857 | 0.32 |
CTB-113P19.1 |
|
3748 |
0.18 |
| chr1_236146948_236147099 | 0.32 |
ENSG00000206803 |
. |
68308 |
0.09 |
| chr12_108292570_108292721 | 0.32 |
ENSG00000263632 |
. |
99148 |
0.07 |
| chr6_132259402_132259553 | 0.32 |
RP11-69I8.3 |
|
12609 |
0.21 |
| chr5_82670520_82670671 | 0.32 |
VCAN |
versican |
96689 |
0.09 |
| chr2_45346900_45347051 | 0.32 |
SIX2 |
SIX homeobox 2 |
110406 |
0.07 |
| chr18_43309151_43309302 | 0.32 |
SLC14A1 |
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
232 |
0.93 |
| chr5_34487773_34488055 | 0.32 |
RAI14 |
retinoic acid induced 14 |
168428 |
0.03 |
| chr3_174978584_174978880 | 0.32 |
NAALADL2-AS2 |
NAALADL2 antisense RNA 2 |
10153 |
0.29 |
| chr20_44548690_44548841 | 0.32 |
PLTP |
phospholipid transfer protein |
7971 |
0.09 |
| chr5_64500241_64500433 | 0.32 |
ENSG00000207439 |
. |
81141 |
0.11 |
| chr9_16706324_16706475 | 0.32 |
BNC2 |
basonuclin 2 |
1319 |
0.52 |
| chr3_134079354_134079505 | 0.32 |
AMOTL2 |
angiomotin like 2 |
11325 |
0.22 |
| chr18_400089_400498 | 0.32 |
RP11-720L2.2 |
|
24123 |
0.21 |
| chr2_53803598_53803749 | 0.32 |
ENSG00000207456 |
. |
6056 |
0.28 |
| chr2_37699858_37700009 | 0.32 |
ENSG00000253078 |
. |
37182 |
0.18 |
| chr8_22860932_22861110 | 0.32 |
PEBP4 |
phosphatidylethanolamine-binding protein 4 |
3508 |
0.15 |
| chr9_113606609_113606760 | 0.32 |
ENSG00000206923 |
. |
58933 |
0.13 |
| chr22_36727795_36728637 | 0.32 |
ENSG00000266345 |
. |
15970 |
0.18 |
| chr1_36173339_36173885 | 0.32 |
ENSG00000239859 |
. |
1737 |
0.31 |
| chr5_9523779_9523930 | 0.32 |
CTD-2201E9.2 |
|
564 |
0.77 |
| chr12_58759641_58759792 | 0.32 |
RP11-362K2.2 |
Protein LOC100506869 |
178191 |
0.03 |
| chr12_2357520_2357671 | 0.32 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
21347 |
0.21 |
| chr18_43308457_43308608 | 0.32 |
SLC14A1 |
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
462 |
0.82 |
| chr4_77532872_77533023 | 0.32 |
AC107072.2 |
|
25829 |
0.15 |
| chr5_111165116_111165267 | 0.31 |
NREP |
neuronal regeneration related protein |
71276 |
0.1 |
| chr7_107284240_107284391 | 0.31 |
SLC26A4 |
solute carrier family 26 (anion exchanger), member 4 |
16765 |
0.14 |
| chr13_32340647_32340798 | 0.31 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
27043 |
0.26 |
| chr13_43579931_43580082 | 0.31 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
13621 |
0.24 |
| chr18_447628_447904 | 0.31 |
RP11-720L2.2 |
|
23350 |
0.19 |
| chr15_70615447_70615598 | 0.31 |
ENSG00000200216 |
. |
129947 |
0.06 |
| chr18_74789491_74789956 | 0.31 |
MBP |
myelin basic protein |
27494 |
0.24 |
| chr22_30440835_30440986 | 0.31 |
HORMAD2 |
HORMA domain containing 2 |
35253 |
0.13 |
| chr5_64493902_64494516 | 0.31 |
ENSG00000207439 |
. |
75013 |
0.12 |
| chr3_123378808_123378959 | 0.31 |
MYLK-AS2 |
MYLK antisense RNA 2 |
29608 |
0.15 |
| chr4_17069080_17069231 | 0.31 |
LDB2 |
LIM domain binding 2 |
168723 |
0.04 |
| chr11_125984167_125984417 | 0.31 |
CDON |
cell adhesion associated, oncogene regulated |
51062 |
0.1 |
| chr20_37602831_37602982 | 0.31 |
DHX35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
11919 |
0.19 |
| chr5_58460603_58460754 | 0.31 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
111267 |
0.07 |
| chr15_70779488_70780159 | 0.31 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
214797 |
0.02 |
| chr1_203167082_203167289 | 0.31 |
CHI3L1 |
chitinase 3-like 1 (cartilage glycoprotein-39) |
11308 |
0.14 |
| chr9_82231282_82231433 | 0.31 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
36151 |
0.25 |
| chr13_77304738_77304889 | 0.31 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
155712 |
0.04 |
| chr20_16904101_16904252 | 0.31 |
ENSG00000212165 |
. |
59502 |
0.15 |
| chr4_151151399_151151550 | 0.31 |
ENSG00000238721 |
. |
20616 |
0.25 |
| chr1_101200153_101200304 | 0.30 |
VCAM1 |
vascular cell adhesion molecule 1 |
14908 |
0.26 |
| chr13_24561197_24561348 | 0.30 |
SPATA13 |
spermatogenesis associated 13 |
7328 |
0.23 |
| chr12_49206263_49206414 | 0.30 |
CACNB3 |
calcium channel, voltage-dependent, beta 3 subunit |
1896 |
0.18 |
| chr1_97024047_97024198 | 0.30 |
ENSG00000241992 |
. |
24643 |
0.26 |
| chr10_63223643_63223794 | 0.30 |
TMEM26 |
transmembrane protein 26 |
10510 |
0.24 |
| chr12_26304780_26304931 | 0.30 |
SSPN |
sarcospan |
16955 |
0.18 |
| chr4_187544517_187544858 | 0.30 |
FAT1 |
FAT atypical cadherin 1 |
25509 |
0.2 |
| chr5_137566220_137566371 | 0.30 |
CDC23 |
cell division cycle 23 |
17263 |
0.12 |
| chr8_110414330_110414481 | 0.30 |
PKHD1L1 |
polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1 |
39699 |
0.16 |
| chr17_71285096_71285247 | 0.30 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
22376 |
0.14 |
| chr6_1614767_1615035 | 0.30 |
FOXC1 |
forkhead box C1 |
4220 |
0.35 |
| chr6_52383740_52383891 | 0.30 |
TRAM2 |
translocation associated membrane protein 2 |
57898 |
0.13 |
| chr5_125473495_125473784 | 0.30 |
ENSG00000265637 |
. |
58889 |
0.16 |
| chr10_128569188_128569339 | 0.30 |
DOCK1 |
dedicator of cytokinesis 1 |
24715 |
0.21 |
| chr4_48390789_48390940 | 0.30 |
SLAIN2 |
SLAIN motif family, member 2 |
5169 |
0.28 |
| chr5_36300674_36300825 | 0.30 |
RANBP3L |
RAN binding protein 3-like |
1253 |
0.53 |
| chr8_105352335_105352486 | 0.30 |
DCSTAMP |
dendrocyte expressed seven transmembrane protein |
356 |
0.9 |
| chr3_114136877_114137028 | 0.30 |
ZBTB20-AS1 |
ZBTB20 antisense RNA 1 |
30436 |
0.18 |
| chr5_34489512_34489663 | 0.30 |
RAI14 |
retinoic acid induced 14 |
166755 |
0.03 |
| chr17_2082426_2082658 | 0.30 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
6070 |
0.13 |
| chr15_57436408_57436559 | 0.30 |
TCF12 |
transcription factor 12 |
75145 |
0.11 |
| chr11_70261652_70262194 | 0.30 |
CTTN |
cortactin |
4308 |
0.17 |
| chr5_121417331_121417482 | 0.29 |
LOX |
lysyl oxidase |
3426 |
0.28 |
| chr4_144318381_144318532 | 0.29 |
GAB1 |
GRB2-associated binding protein 1 |
5793 |
0.26 |
| chr20_18685450_18685672 | 0.29 |
RP11-379J5.5 |
|
6208 |
0.24 |
| chr12_49653469_49653759 | 0.29 |
RP11-977B10.2 |
|
4925 |
0.12 |
| chr2_121695887_121696038 | 0.29 |
ENSG00000221321 |
. |
96687 |
0.08 |
| chr18_53869860_53870011 | 0.29 |
ENSG00000201816 |
. |
123110 |
0.06 |
| chr7_51030538_51030756 | 0.29 |
ENSG00000201133 |
. |
27457 |
0.23 |
| chr12_64773878_64774029 | 0.29 |
C12orf56 |
chromosome 12 open reading frame 56 |
10353 |
0.17 |
| chr12_63253482_63253831 | 0.29 |
ENSG00000200296 |
. |
8975 |
0.26 |
| chr15_49966362_49966513 | 0.29 |
RP11-353B9.1 |
|
22101 |
0.2 |
| chr9_113506622_113506773 | 0.29 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
30930 |
0.23 |
| chr10_22542450_22542989 | 0.29 |
EBLN1 |
endogenous Bornavirus-like nucleoprotein 1 |
43769 |
0.13 |
| chr9_4838397_4838548 | 0.29 |
RCL1 |
RNA terminal phosphate cyclase-like 1 |
1046 |
0.47 |
| chr8_12978290_12978441 | 0.29 |
DLC1 |
deleted in liver cancer 1 |
4612 |
0.27 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.1 | 0.3 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.3 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.1 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.2 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.1 | 0.4 | GO:0015840 | urea transport(GO:0015840) |
| 0.1 | 0.3 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.3 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.3 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.1 | GO:0072191 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.1 | 0.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0061043 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.2 | GO:0061525 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.4 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.2 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0048342 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.3 | GO:0050922 | negative regulation of chemotaxis(GO:0050922) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.4 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.0 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.2 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0046325 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:2000846 | corticosteroid hormone secretion(GO:0035930) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) |
| 0.0 | 0.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.2 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.3 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.3 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.0 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0060592 | mammary gland formation(GO:0060592) mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0071027 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:1902170 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:0060462 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0060412 | ventricular septum morphogenesis(GO:0060412) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:0051938 | L-glutamate import(GO:0051938) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.0 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.0 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.0 | GO:0070091 | glucagon secretion(GO:0070091) |
| 0.0 | 0.0 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.0 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.0 | GO:1903020 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0060872 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) urinary bladder development(GO:0060157) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process(GO:0009147) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0072243 | metanephric nephron tubule development(GO:0072234) metanephric nephron epithelium development(GO:0072243) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.0 | GO:0016241 | regulation of macroautophagy(GO:0016241) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0009130 | UMP biosynthetic process(GO:0006222) pyrimidine nucleoside monophosphate biosynthetic process(GO:0009130) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0032069 | regulation of nuclease activity(GO:0032069) |
| 0.0 | 0.0 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.2 | GO:0006195 | purine nucleotide catabolic process(GO:0006195) |
| 0.0 | 0.1 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.0 | GO:0051883 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0071450 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.0 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.0 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.0 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 2.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 1.0 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.4 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0030354 | melanin-concentrating hormone activity(GO:0030354) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 0.1 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |