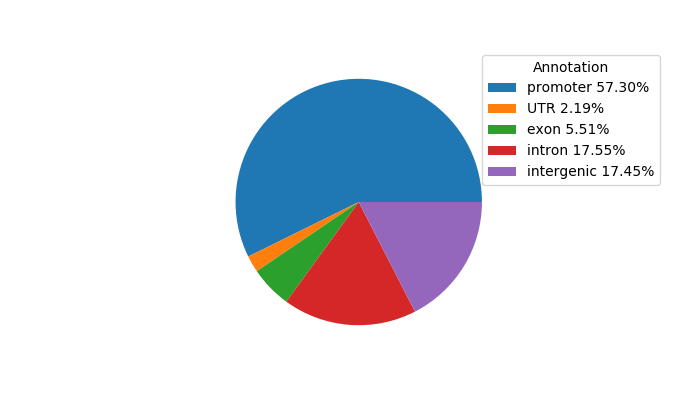
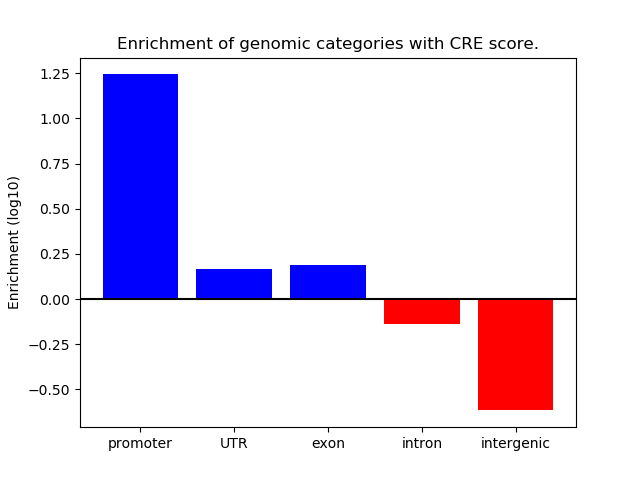
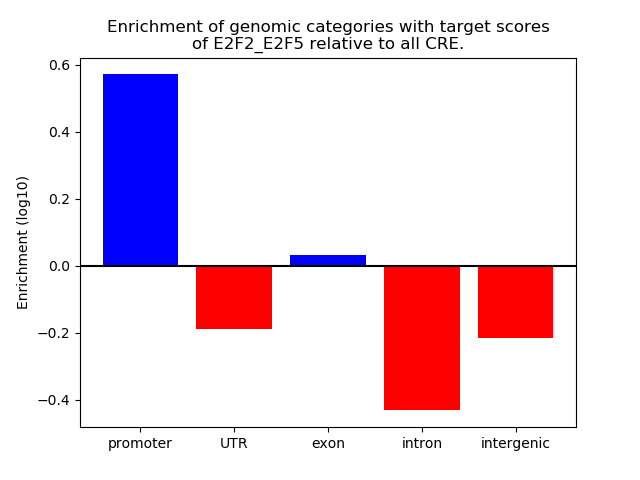
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
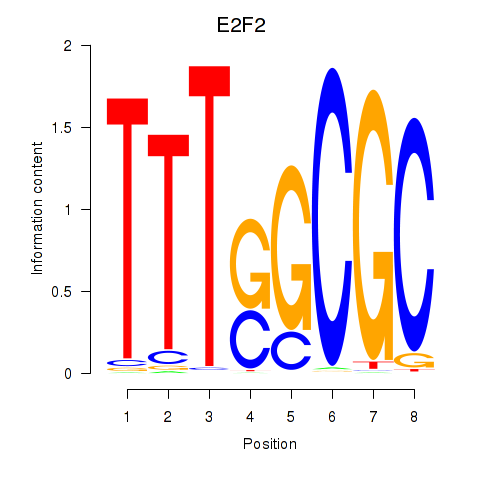
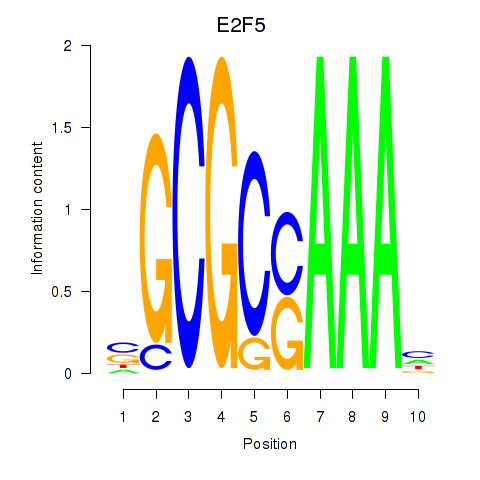
Results for E2F2_E2F5
Z-value: 1.74
Transcription factors associated with E2F2_E2F5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F2
|
ENSG00000007968.6 | E2F transcription factor 2 |
|
E2F5
|
ENSG00000133740.6 | E2F transcription factor 5 |
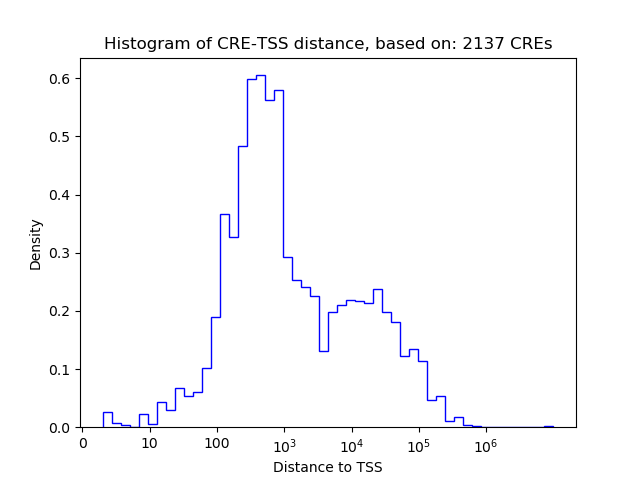
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_23866536_23866738 | E2F2 | 8925 | 0.175245 | 0.88 | 1.9e-03 | Click! |
| chr1_23857278_23857429 | E2F2 | 359 | 0.864837 | 0.74 | 2.4e-02 | Click! |
| chr1_23857552_23857703 | E2F2 | 85 | 0.969441 | 0.72 | 3.0e-02 | Click! |
| chr1_23866295_23866462 | E2F2 | 8666 | 0.176019 | 0.70 | 3.5e-02 | Click! |
| chr1_23856635_23856974 | E2F2 | 908 | 0.568191 | 0.69 | 3.8e-02 | Click! |
| chr8_86113087_86113275 | E2F5 | 5217 | 0.141711 | -0.57 | 1.1e-01 | Click! |
| chr8_86102063_86102214 | E2F5 | 2218 | 0.218893 | 0.29 | 4.5e-01 | Click! |
| chr8_86101250_86101401 | E2F5 | 1405 | 0.323320 | 0.18 | 6.4e-01 | Click! |
| chr8_86104958_86105109 | E2F5 | 5113 | 0.145131 | 0.13 | 7.3e-01 | Click! |
Activity of the E2F2_E2F5 motif across conditions
Conditions sorted by the z-value of the E2F2_E2F5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
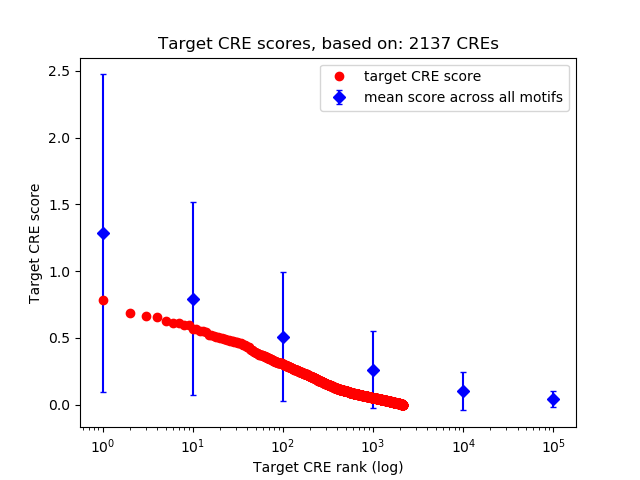
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_16474453_16474697 | 0.78 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
1811 |
0.29 |
| chr1_12201058_12201219 | 0.69 |
TNFRSF8 |
tumor necrosis factor receptor superfamily, member 8 |
15182 |
0.15 |
| chr1_205290600_205290751 | 0.66 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
208 |
0.93 |
| chr4_40057808_40058129 | 0.66 |
N4BP2 |
NEDD4 binding protein 2 |
478 |
0.8 |
| chr17_58678203_58678354 | 0.63 |
PPM1D |
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
734 |
0.66 |
| chr4_1685292_1685462 | 0.61 |
ENSG00000207009 |
. |
121 |
0.76 |
| chr19_37287796_37287970 | 0.61 |
CTD-2162K18.5 |
|
537 |
0.7 |
| chr3_153839356_153839507 | 0.60 |
ARHGEF26 |
Rho guanine nucleotide exchange factor (GEF) 26 |
202 |
0.64 |
| chr21_34914620_34914771 | 0.60 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
26 |
0.74 |
| chr2_175350906_175351070 | 0.57 |
GPR155 |
G protein-coupled receptor 155 |
777 |
0.66 |
| chr5_69321527_69321686 | 0.57 |
SERF1B |
small EDRK-rich factor 1B (centromeric) |
245 |
0.94 |
| chr15_74907592_74907743 | 0.55 |
CTD-3154N5.1 |
|
270 |
0.55 |
| chr4_108911771_108911922 | 0.55 |
HADH |
hydroxyacyl-CoA dehydrogenase |
788 |
0.64 |
| chr13_50070457_50070608 | 0.55 |
PHF11 |
PHD finger protein 11 |
1 |
0.98 |
| chrX_11683937_11684088 | 0.52 |
ARHGAP6 |
Rho GTPase activating protein 6 |
191 |
0.97 |
| chr5_107006410_107006592 | 0.52 |
EFNA5 |
ephrin-A5 |
95 |
0.98 |
| chr1_235247512_235247663 | 0.52 |
ENSG00000207181 |
. |
43665 |
0.13 |
| chr7_100034344_100034495 | 0.51 |
PPP1R35 |
protein phosphatase 1, regulatory subunit 35 |
231 |
0.69 |
| chr15_66995452_66995652 | 0.51 |
SMAD6 |
SMAD family member 6 |
878 |
0.68 |
| chr10_25013084_25013502 | 0.50 |
ARHGAP21 |
Rho GTPase activating protein 21 |
696 |
0.79 |
| chr6_150039448_150039648 | 0.50 |
LATS1 |
large tumor suppressor kinase 1 |
156 |
0.94 |
| chr17_78966391_78966691 | 0.49 |
CHMP6 |
charged multivesicular body protein 6 |
900 |
0.43 |
| chr2_233180899_233181095 | 0.49 |
DIS3L2 |
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
17656 |
0.17 |
| chr7_76139277_76139433 | 0.49 |
UPK3B |
uroplakin 3B |
390 |
0.81 |
| chr11_57123702_57124132 | 0.48 |
ENSG00000266018 |
. |
3279 |
0.12 |
| chr2_219575773_219575924 | 0.48 |
TTLL4 |
tubulin tyrosine ligase-like family, member 4 |
131 |
0.94 |
| chr14_24665331_24665657 | 0.48 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
601 |
0.42 |
| chr9_137393142_137393331 | 0.48 |
RXRA |
retinoid X receptor, alpha |
94808 |
0.07 |
| chr11_106889622_106889773 | 0.47 |
GUCY1A2 |
guanylate cyclase 1, soluble, alpha 2 |
447 |
0.91 |
| chr15_47476999_47477319 | 0.47 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
701 |
0.73 |
| chr15_43415038_43415301 | 0.47 |
TMEM62 |
transmembrane protein 62 |
308 |
0.89 |
| chr15_71184789_71184970 | 0.46 |
LRRC49 |
leucine rich repeat containing 49 |
86 |
0.56 |
| chr2_242625596_242625805 | 0.46 |
DTYMK |
deoxythymidylate kinase (thymidylate kinase) |
408 |
0.72 |
| chr12_21810059_21810237 | 0.46 |
LDHB |
lactate dehydrogenase B |
148 |
0.96 |
| chr2_105473259_105473410 | 0.46 |
POU3F3 |
POU class 3 homeobox 3 |
1365 |
0.32 |
| chr18_37379955_37380106 | 0.45 |
ENSG00000212354 |
. |
121444 |
0.06 |
| chr2_175351124_175351275 | 0.45 |
GPR155 |
G protein-coupled receptor 155 |
566 |
0.77 |
| chr12_122984418_122984836 | 0.45 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
168 |
0.94 |
| chr11_67165283_67165521 | 0.44 |
ENSG00000253024 |
. |
2620 |
0.1 |
| chr14_95234863_95235014 | 0.44 |
GSC |
goosecoid homeobox |
1624 |
0.5 |
| chr8_98289888_98290039 | 0.43 |
TSPYL5 |
TSPY-like 5 |
213 |
0.96 |
| chr7_143599335_143599486 | 0.43 |
FAM115A |
family with sequence similarity 115, member A |
119 |
0.96 |
| chr2_172380242_172380480 | 0.42 |
CYBRD1 |
cytochrome b reductase 1 |
772 |
0.73 |
| chr2_148394331_148394482 | 0.41 |
ENSG00000253083 |
. |
76717 |
0.11 |
| chr5_41870001_41870238 | 0.41 |
ENSG00000248668 |
. |
13 |
0.91 |
| chr19_18530764_18530945 | 0.41 |
SSBP4 |
single stranded DNA binding protein 4 |
59 |
0.94 |
| chr2_91997601_91997765 | 0.40 |
IGKV1OR-1 |
immunoglobulin kappa variable 1/OR-1 (pseudogene) |
8114 |
0.25 |
| chr2_27631816_27631967 | 0.40 |
PPM1G |
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
558 |
0.58 |
| chr4_151937156_151937307 | 0.40 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
352 |
0.89 |
| chr19_50353966_50354117 | 0.39 |
PTOV1 |
prostate tumor overexpressed 1 |
30 |
0.75 |
| chr19_30432531_30432893 | 0.39 |
URI1 |
URI1, prefoldin-like chaperone |
446 |
0.9 |
| chr8_94752567_94752718 | 0.39 |
RBM12B-AS1 |
RBM12B antisense RNA 1 |
293 |
0.43 |
| chr7_90226220_90226410 | 0.38 |
AC002456.2 |
|
352 |
0.61 |
| chr3_50377830_50377981 | 0.38 |
RASSF1 |
Ras association (RalGDS/AF-6) domain family member 1 |
367 |
0.53 |
| chr5_150473994_150474160 | 0.38 |
TNIP1 |
TNFAIP3 interacting protein 1 |
939 |
0.58 |
| chr4_55096263_55096414 | 0.37 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
151 |
0.97 |
| chr2_221969094_221969245 | 0.37 |
EPHA4 |
EPH receptor A4 |
398111 |
0.01 |
| chr7_99716711_99716862 | 0.37 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
141 |
0.74 |
| chr11_65687460_65687611 | 0.37 |
DRAP1 |
DR1-associated protein 1 (negative cofactor 2 alpha) |
340 |
0.67 |
| chr3_72127692_72127843 | 0.37 |
ENSG00000212070 |
. |
183812 |
0.03 |
| chr13_46755573_46755724 | 0.37 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
811 |
0.58 |
| chr10_70320973_70321232 | 0.36 |
TET1 |
tet methylcytosine dioxygenase 1 |
689 |
0.69 |
| chr7_27204759_27204916 | 0.36 |
HOXA9 |
homeobox A9 |
308 |
0.69 |
| chr3_136824240_136824391 | 0.36 |
IL20RB-AS1 |
IL20RB antisense RNA 1 |
123277 |
0.06 |
| chr8_27695395_27695562 | 0.36 |
PBK |
PDZ binding kinase |
121 |
0.96 |
| chr12_49374639_49374845 | 0.35 |
WNT1 |
wingless-type MMTV integration site family, member 1 |
2344 |
0.11 |
| chr18_11850127_11850406 | 0.35 |
CHMP1B |
charged multivesicular body protein 1B |
1129 |
0.36 |
| chr5_271481_271632 | 0.35 |
CTD-2083E4.6 |
|
75 |
0.65 |
| chr8_22551640_22551890 | 0.35 |
EGR3 |
early growth response 3 |
950 |
0.44 |
| chr1_31769225_31769468 | 0.35 |
SNRNP40 |
small nuclear ribonucleoprotein 40kDa (U5) |
271 |
0.69 |
| chr11_58874195_58874346 | 0.35 |
FAM111B |
family with sequence similarity 111, member B |
388 |
0.85 |
| chr12_4381658_4381881 | 0.35 |
CCND2 |
cyclin D2 |
1169 |
0.41 |
| chr19_52773021_52773172 | 0.34 |
ZNF766 |
zinc finger protein 766 |
250 |
0.85 |
| chr10_62703240_62703501 | 0.34 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
635 |
0.8 |
| chr17_35292034_35292185 | 0.33 |
RP11-445F12.1 |
|
1812 |
0.23 |
| chr10_11059547_11059708 | 0.33 |
CELF2 |
CUGBP, Elav-like family member 2 |
266 |
0.92 |
| chr11_62541978_62542240 | 0.33 |
TAF6L |
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
2598 |
0.08 |
| chr3_55522055_55522206 | 0.33 |
WNT5A-AS1 |
WNT5A antisense RNA 1 |
403 |
0.72 |
| chr6_52929743_52930148 | 0.33 |
FBXO9 |
F-box protein 9 |
149 |
0.93 |
| chrX_132092122_132092387 | 0.33 |
HS6ST2 |
heparan sulfate 6-O-sulfotransferase 2 |
910 |
0.71 |
| chr6_106328518_106328778 | 0.33 |
ENSG00000200198 |
. |
23600 |
0.27 |
| chrX_55515914_55516065 | 0.32 |
USP51 |
ubiquitin specific peptidase 51 |
354 |
0.89 |
| chrX_73164517_73164668 | 0.32 |
RP13-216E22.5 |
|
24728 |
0.23 |
| chr17_7900691_7900842 | 0.32 |
GUCY2D |
guanylate cyclase 2D, membrane (retina-specific) |
5146 |
0.12 |
| chr3_169720111_169720354 | 0.32 |
SEC62-AS1 |
SEC62 antisense RNA 1 |
16729 |
0.13 |
| chr19_14318195_14318346 | 0.32 |
LPHN1 |
latrophilin 1 |
1271 |
0.38 |
| chr11_44748743_44748894 | 0.32 |
TSPAN18 |
tetraspanin 18 |
94 |
0.98 |
| chr19_40315056_40315207 | 0.32 |
DYRK1B |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
7376 |
0.16 |
| chr6_3229015_3229236 | 0.31 |
TUBB2B |
tubulin, beta 2B class IIb |
1156 |
0.44 |
| chr6_8102779_8102930 | 0.31 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
43 |
0.98 |
| chr19_16946806_16946957 | 0.31 |
SIN3B |
SIN3 transcription regulator family member B |
5882 |
0.14 |
| chr11_118272145_118272296 | 0.31 |
ATP5L |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
127 |
0.77 |
| chr22_39993919_39994070 | 0.31 |
CACNA1I |
calcium channel, voltage-dependent, T type, alpha 1I subunit |
27236 |
0.17 |
| chr11_58673004_58673248 | 0.31 |
AP001652.1 |
Uncharacterized protein; cDNA FLJ60524 |
229 |
0.91 |
| chr20_2082194_2082410 | 0.31 |
STK35 |
serine/threonine kinase 35 |
45 |
0.98 |
| chr2_232573942_232574135 | 0.31 |
PTMA |
prothymosin, alpha |
453 |
0.78 |
| chr1_243431690_243431841 | 0.31 |
SDCCAG8 |
serologically defined colon cancer antigen 8 |
12393 |
0.18 |
| chr12_57481350_57481630 | 0.30 |
NAB2 |
NGFI-A binding protein 2 (EGR1 binding protein 2) |
1187 |
0.3 |
| chr10_11652685_11653021 | 0.30 |
RP11-138I18.1 |
|
451 |
0.7 |
| chr1_117664616_117664767 | 0.30 |
TRIM45 |
tripartite motif containing 45 |
277 |
0.9 |
| chr2_19558652_19558898 | 0.30 |
OSR1 |
odd-skipped related transciption factor 1 |
361 |
0.91 |
| chr9_23831341_23831632 | 0.30 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
5151 |
0.37 |
| chr3_183498240_183498391 | 0.30 |
YEATS2 |
YEATS domain containing 2 |
4536 |
0.17 |
| chr10_131935434_131935764 | 0.30 |
GLRX3 |
glutaredoxin 3 |
936 |
0.73 |
| chr5_43064735_43064935 | 0.30 |
CTD-2201E18.3 |
|
2686 |
0.21 |
| chr14_97879326_97879526 | 0.30 |
ENSG00000240730 |
. |
117084 |
0.07 |
| chr17_25782767_25783007 | 0.29 |
KSR1 |
kinase suppressor of ras 1 |
783 |
0.65 |
| chr16_70834690_70834841 | 0.29 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
299 |
0.89 |
| chr7_54732618_54732769 | 0.29 |
ENSG00000200471 |
. |
43975 |
0.16 |
| chr3_4509025_4509208 | 0.29 |
SUMF1 |
sulfatase modifying factor 1 |
151 |
0.96 |
| chr15_93180047_93180198 | 0.29 |
FAM174B |
family with sequence similarity 174, member B |
2352 |
0.36 |
| chr4_74486561_74486712 | 0.29 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
288 |
0.94 |
| chr4_174256659_174256871 | 0.29 |
HMGB2 |
high mobility group box 2 |
489 |
0.74 |
| chr14_64400542_64400693 | 0.29 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
24801 |
0.22 |
| chr16_70729532_70729806 | 0.29 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
173 |
0.93 |
| chr2_105275814_105276048 | 0.29 |
ENSG00000207249 |
. |
73761 |
0.1 |
| chr4_39045763_39045975 | 0.29 |
KLHL5 |
kelch-like family member 5 |
790 |
0.66 |
| chr21_28216861_28217012 | 0.28 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
792 |
0.73 |
| chr2_26681881_26682032 | 0.28 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
10789 |
0.2 |
| chr12_12877982_12878133 | 0.28 |
APOLD1 |
apolipoprotein L domain containing 1 |
808 |
0.43 |
| chr10_108758695_108758846 | 0.28 |
ENSG00000200626 |
. |
28991 |
0.26 |
| chr2_113190370_113190521 | 0.28 |
RGPD8 |
RANBP2-like and GRIP domain containing 8 |
651 |
0.76 |
| chrX_39865999_39866213 | 0.28 |
BCOR |
BCL6 corepressor |
56084 |
0.17 |
| chr9_104500822_104500973 | 0.27 |
GRIN3A |
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A |
35 |
0.99 |
| chr15_45405087_45405238 | 0.27 |
DUOX2 |
dual oxidase 2 |
1197 |
0.25 |
| chr3_32822211_32822366 | 0.27 |
TRIM71 |
tripartite motif containing 71, E3 ubiquitin protein ligase |
37222 |
0.15 |
| chr1_27682961_27683240 | 0.27 |
MAP3K6 |
mitogen-activated protein kinase kinase kinase 6 |
138 |
0.93 |
| chr14_99009042_99009193 | 0.27 |
C14orf177 |
chromosome 14 open reading frame 177 |
168833 |
0.04 |
| chr9_84305434_84305645 | 0.27 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
1319 |
0.53 |
| chr12_31811913_31812064 | 0.27 |
METTL20 |
methyltransferase like 20 |
133 |
0.96 |
| chr11_17374066_17374428 | 0.27 |
NCR3LG1 |
natural killer cell cytotoxicity receptor 3 ligand 1 |
974 |
0.52 |
| chr3_14219692_14219843 | 0.27 |
LSM3 |
LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
91 |
0.77 |
| chr10_99051887_99052112 | 0.26 |
ARHGAP19-SLIT1 |
ARHGAP19-SLIT1 readthrough (NMD candidate) |
385 |
0.46 |
| chr2_47403393_47403544 | 0.26 |
CALM2 |
calmodulin 2 (phosphorylase kinase, delta) |
182 |
0.96 |
| chr1_23884944_23885095 | 0.26 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
1266 |
0.44 |
| chr2_190648545_190648696 | 0.26 |
ORMDL1 |
ORM1-like 1 (S. cerevisiae) |
321 |
0.59 |
| chr16_3550718_3550869 | 0.26 |
CLUAP1 |
clusterin associated protein 1 |
131 |
0.94 |
| chr6_142467659_142468075 | 0.26 |
VTA1 |
vesicle (multivesicular body) trafficking 1 |
500 |
0.81 |
| chr15_44719994_44720235 | 0.26 |
CTDSPL2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
102 |
0.91 |
| chr16_68563969_68564120 | 0.26 |
ZFP90 |
ZFP90 zinc finger protein |
7 |
0.97 |
| chr8_25903900_25904102 | 0.26 |
EBF2 |
early B-cell factor 2 |
1088 |
0.66 |
| chrX_118753397_118753600 | 0.26 |
NKRF |
NFKB repressing factor |
13640 |
0.15 |
| chr5_136899668_136899823 | 0.26 |
ENSG00000221612 |
. |
30602 |
0.19 |
| chr20_35724684_35724840 | 0.26 |
RBL1 |
retinoblastoma-like 1 (p107) |
364 |
0.9 |
| chr4_90221268_90221504 | 0.26 |
GPRIN3 |
GPRIN family member 3 |
7775 |
0.31 |
| chr2_46526159_46526310 | 0.26 |
EPAS1 |
endothelial PAS domain protein 1 |
1693 |
0.49 |
| chr10_125846427_125846578 | 0.25 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
5468 |
0.31 |
| chr12_94853936_94854087 | 0.25 |
CCDC41 |
coiled-coil domain containing 41 |
247 |
0.94 |
| chr12_58223196_58223436 | 0.25 |
CTDSP2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
17 |
0.94 |
| chr12_95868302_95868453 | 0.25 |
METAP2 |
methionyl aminopeptidase 2 |
439 |
0.84 |
| chr10_129861675_129861865 | 0.25 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
15936 |
0.26 |
| chr17_4607619_4607770 | 0.25 |
PELP1 |
proline, glutamate and leucine rich protein 1 |
62 |
0.63 |
| chr3_107352295_107352521 | 0.25 |
BBX |
bobby sox homolog (Drosophila) |
12275 |
0.31 |
| chr15_47476608_47476882 | 0.25 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
447 |
0.85 |
| chr1_35908347_35908498 | 0.24 |
KIAA0319L |
KIAA0319-like |
11886 |
0.17 |
| chr1_22432425_22432576 | 0.24 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
23844 |
0.15 |
| chr14_52959290_52959441 | 0.24 |
TXNDC16 |
thioredoxin domain containing 16 |
59858 |
0.12 |
| chr2_225905663_225905934 | 0.24 |
DOCK10 |
dedicator of cytokinesis 10 |
1361 |
0.55 |
| chr18_61638270_61638435 | 0.24 |
SERPINB8 |
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
613 |
0.73 |
| chr1_198744986_198745208 | 0.24 |
ENSG00000207975 |
. |
83014 |
0.09 |
| chr21_45077557_45077755 | 0.24 |
HSF2BP |
heat shock transcription factor 2 binding protein |
369 |
0.84 |
| chr20_822843_822995 | 0.24 |
FAM110A |
family with sequence similarity 110, member A |
2366 |
0.35 |
| chr11_10329277_10329428 | 0.24 |
ADM |
adrenomedullin |
2428 |
0.23 |
| chr19_57351053_57351204 | 0.24 |
PEG3 |
paternally expressed 3 |
936 |
0.39 |
| chr6_42858705_42858888 | 0.24 |
C6orf226 |
chromosome 6 open reading frame 226 |
242 |
0.88 |
| chr2_113590430_113590581 | 0.24 |
IL1B |
interleukin 1, beta |
3506 |
0.21 |
| chr13_50707168_50707390 | 0.24 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
50972 |
0.13 |
| chr3_141030206_141030479 | 0.24 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
12713 |
0.22 |
| chr15_53080371_53080530 | 0.24 |
ONECUT1 |
one cut homeobox 1 |
1759 |
0.48 |
| chr1_54665245_54665442 | 0.23 |
CYB5RL |
cytochrome b5 reductase-like |
342 |
0.59 |
| chr16_84401523_84401674 | 0.23 |
ATP2C2 |
ATPase, Ca++ transporting, type 2C, member 2 |
535 |
0.79 |
| chr10_81850627_81850778 | 0.23 |
TMEM254 |
transmembrane protein 254 |
9223 |
0.18 |
| chr8_143859283_143859453 | 0.23 |
LYNX1 |
Ly6/neurotoxin 1 |
142 |
0.91 |
| chr1_26362139_26362300 | 0.23 |
SLC30A2 |
solute carrier family 30 (zinc transporter), member 2 |
10385 |
0.1 |
| chr22_50634186_50634337 | 0.23 |
TRABD |
TraB domain containing |
2818 |
0.11 |
| chr15_89949515_89949757 | 0.23 |
ENSG00000207819 |
. |
38388 |
0.14 |
| chr21_45138541_45138692 | 0.23 |
PDXK |
pyridoxal (pyridoxine, vitamin B6) kinase |
359 |
0.88 |
| chr14_24610403_24610554 | 0.23 |
EMC9 |
ER membrane protein complex subunit 9 |
301 |
0.7 |
| chr2_177502756_177502963 | 0.23 |
ENSG00000252027 |
. |
26545 |
0.25 |
| chr4_1161143_1161294 | 0.23 |
RP11-20I20.4 |
|
348 |
0.81 |
| chr15_56756676_56757156 | 0.23 |
MNS1 |
meiosis-specific nuclear structural 1 |
419 |
0.9 |
| chr1_101701824_101702062 | 0.23 |
RP4-575N6.4 |
|
141 |
0.83 |
| chr17_74380229_74380402 | 0.23 |
PRPSAP1 |
phosphoribosyl pyrophosphate synthetase-associated protein 1 |
250 |
0.55 |
| chr6_26088017_26088347 | 0.23 |
HFE |
hemochromatosis |
515 |
0.56 |
| chr8_81806183_81806334 | 0.23 |
ZNF704 |
zinc finger protein 704 |
19242 |
0.26 |
| chr19_50935443_50935594 | 0.22 |
MYBPC2 |
myosin binding protein C, fast type |
642 |
0.51 |
| chr11_13300608_13301500 | 0.22 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
1622 |
0.51 |
| chrX_129090519_129090731 | 0.22 |
RP4-537K23.4 |
|
879 |
0.61 |
| chr7_27162728_27162879 | 0.22 |
HOXA-AS2 |
HOXA cluster antisense RNA 2 |
16 |
0.93 |
| chr1_149982777_149982991 | 0.22 |
OTUD7B |
OTU domain containing 7B |
259 |
0.9 |
| chr13_31735240_31735730 | 0.22 |
HSPH1 |
heat shock 105kDa/110kDa protein 1 |
571 |
0.83 |
| chr9_77112695_77112868 | 0.22 |
RORB |
RAR-related orphan receptor B |
500 |
0.75 |
| chr19_4519048_4519199 | 0.22 |
PLIN4 |
perilipin 4 |
1407 |
0.24 |
| chr19_45490552_45490703 | 0.22 |
CLPTM1 |
cleft lip and palate associated transmembrane protein 1 |
4899 |
0.1 |
| chr1_33190488_33190689 | 0.22 |
RP11-114B7.6 |
|
250 |
0.9 |
| chr17_46048605_46048756 | 0.22 |
CDK5RAP3 |
CDK5 regulatory subunit associated protein 3 |
163 |
0.9 |
| chr6_82461023_82461181 | 0.22 |
FAM46A |
family with sequence similarity 46, member A |
368 |
0.88 |
| chr16_31712181_31712332 | 0.22 |
CLUHP3 |
clustered mitochondria (cluA/CLU1) homolog pseudogene 3 |
2233 |
0.26 |
| chr7_48285350_48285616 | 0.22 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
74426 |
0.12 |
| chr3_42623874_42624044 | 0.22 |
SEC22C |
SEC22 vesicle trafficking protein homolog C (S. cerevisiae) |
140 |
0.86 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 0.4 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.1 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.5 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0072162 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 0.4 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.1 | 0.2 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.2 | GO:0003079 | obsolete positive regulation of natriuresis(GO:0003079) |
| 0.1 | 0.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 0.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.2 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.2 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.6 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.2 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.2 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0051940 | regulation of neurotransmitter uptake(GO:0051580) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.4 | GO:0018200 | peptidyl-glutamic acid modification(GO:0018200) |
| 0.0 | 0.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.0 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.0 | GO:0052305 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) positive regulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052555) positive regulation by symbiont of host immune response(GO:0052556) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:2000192 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.2 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.3 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.1 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.0 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.0 | GO:1901320 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 | 0.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.2 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.0 | 0.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.1 | GO:0098657 | neurotransmitter uptake(GO:0001504) import into cell(GO:0098657) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.3 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.2 | GO:0006862 | nucleotide transport(GO:0006862) |
| 0.0 | 0.1 | GO:0006385 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.4 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.1 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.3 | GO:0043928 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.3 | GO:0051318 | G1 phase(GO:0051318) |
| 0.0 | 0.0 | GO:0010613 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.8 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0042627 | chylomicron(GO:0042627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.5 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.4 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.2 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.4 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.1 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.4 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.3 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.0 | 0.5 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.0 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.0 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |