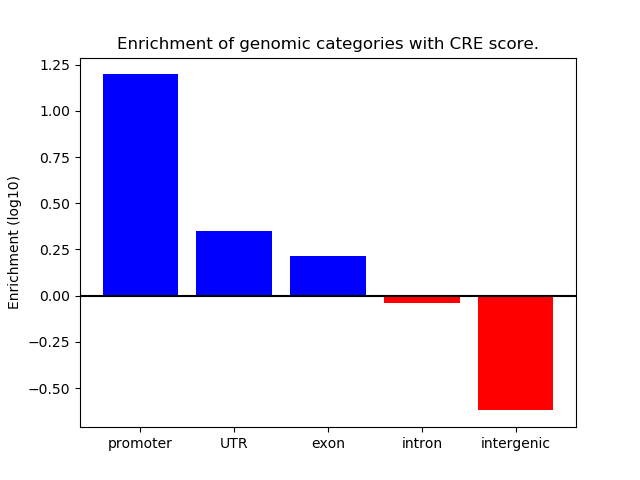
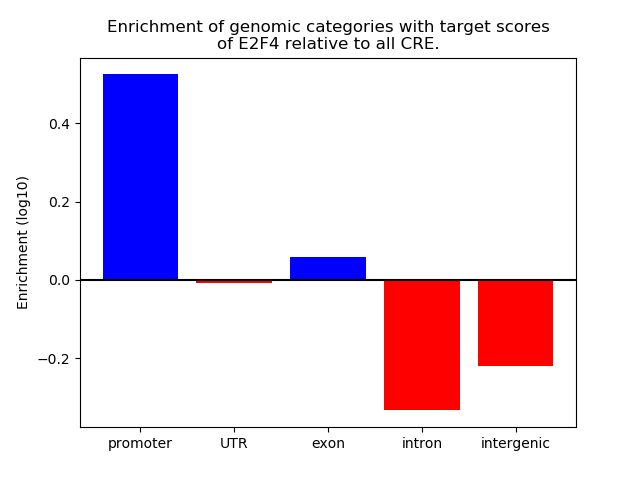
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
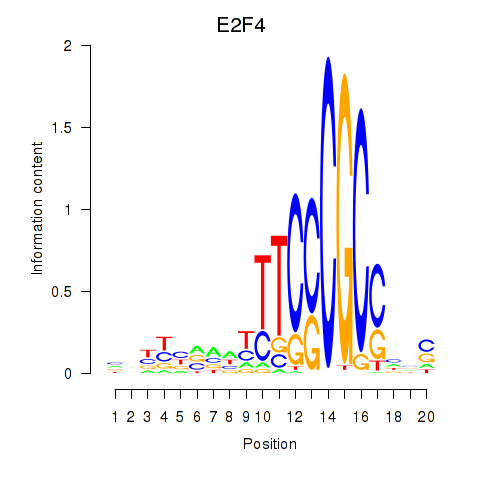
Results for E2F4
Z-value: 1.08
Transcription factors associated with E2F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F4
|
ENSG00000205250.4 | E2F transcription factor 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_67226809_67227108 | E2F4 | 886 | 0.288743 | 0.62 | 7.6e-02 | Click! |
| chr16_67226498_67226740 | E2F4 | 547 | 0.478339 | 0.59 | 9.6e-02 | Click! |
| chr16_67225330_67225481 | E2F4 | 667 | 0.368028 | 0.22 | 5.6e-01 | Click! |
| chr16_67225481_67225632 | E2F4 | 516 | 0.483180 | -0.15 | 7.1e-01 | Click! |
| chr16_67227161_67227492 | E2F4 | 1254 | 0.192017 | 0.14 | 7.2e-01 | Click! |
Activity of the E2F4 motif across conditions
Conditions sorted by the z-value of the E2F4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_37620705_37620902 | 0.72 |
SSTR3 |
somatostatin receptor 3 |
12441 |
0.13 |
| chr8_54794402_54794553 | 0.52 |
RGS20 |
regulator of G-protein signaling 20 |
1023 |
0.52 |
| chr16_68108782_68108933 | 0.49 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
10390 |
0.1 |
| chr19_13044178_13044329 | 0.48 |
FARSA |
phenylalanyl-tRNA synthetase, alpha subunit |
241 |
0.77 |
| chr14_99729801_99729952 | 0.47 |
AL109767.1 |
|
591 |
0.77 |
| chr11_117922812_117923152 | 0.47 |
ENSG00000272075 |
. |
14970 |
0.15 |
| chr3_63848828_63849118 | 0.45 |
THOC7 |
THO complex 7 homolog (Drosophila) |
606 |
0.58 |
| chr9_103114517_103114668 | 0.45 |
TEX10 |
testis expressed 10 |
261 |
0.93 |
| chr22_32341357_32341508 | 0.42 |
C22orf24 |
chromosome 22 open reading frame 24 |
96 |
0.93 |
| chr5_65018218_65018648 | 0.40 |
NLN |
neurolysin (metallopeptidase M3 family) |
363 |
0.54 |
| chr21_34392077_34392228 | 0.39 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
6001 |
0.22 |
| chr15_67359185_67359373 | 0.38 |
SMAD3 |
SMAD family member 3 |
1096 |
0.64 |
| chr2_157293041_157293192 | 0.37 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
119 |
0.97 |
| chr17_61780737_61780956 | 0.36 |
STRADA |
STE20-related kinase adaptor alpha |
969 |
0.46 |
| chr6_42359385_42359536 | 0.35 |
ENSG00000221252 |
. |
20674 |
0.21 |
| chr2_219723652_219723803 | 0.34 |
WNT6 |
wingless-type MMTV integration site family, member 6 |
817 |
0.51 |
| chrX_17674771_17674922 | 0.34 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
21433 |
0.23 |
| chr4_2787896_2788215 | 0.34 |
SH3BP2 |
SH3-domain binding protein 2 |
6695 |
0.21 |
| chr19_40336344_40336545 | 0.33 |
FBL |
fibrillarin |
525 |
0.75 |
| chr18_22042462_22042613 | 0.33 |
HRH4 |
histamine receptor H4 |
1844 |
0.3 |
| chr19_10982774_10983002 | 0.32 |
CARM1 |
coactivator-associated arginine methyltransferase 1 |
509 |
0.7 |
| chr18_12308768_12309026 | 0.32 |
TUBB6 |
tubulin, beta 6 class V |
635 |
0.7 |
| chr14_102666358_102666589 | 0.32 |
WDR20 |
WD repeat domain 20 |
5190 |
0.21 |
| chr2_161349301_161349452 | 0.31 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
653 |
0.81 |
| chr20_5930659_5930947 | 0.31 |
TRMT6 |
tRNA methyltransferase 6 homolog (S. cerevisiae) |
248 |
0.7 |
| chr15_91106977_91107236 | 0.31 |
CRTC3 |
CREB regulated transcription coactivator 3 |
29839 |
0.14 |
| chr15_37392504_37392655 | 0.31 |
MEIS2 |
Meis homeobox 2 |
125 |
0.94 |
| chr2_112237136_112237426 | 0.31 |
ENSG00000266139 |
. |
158613 |
0.04 |
| chr4_13485282_13485564 | 0.31 |
RAB28 |
RAB28, member RAS oncogene family |
566 |
0.83 |
| chr5_171542288_171542655 | 0.30 |
ENSG00000266671 |
. |
34491 |
0.16 |
| chr22_25462879_25463030 | 0.30 |
KIAA1671 |
KIAA1671 |
2982 |
0.31 |
| chr2_203736496_203736695 | 0.30 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
113 |
0.97 |
| chr21_36399444_36399595 | 0.30 |
RUNX1 |
runt-related transcription factor 1 |
21943 |
0.28 |
| chr10_116252774_116252925 | 0.30 |
ABLIM1 |
actin binding LIM protein 1 |
5097 |
0.29 |
| chr20_18448159_18448354 | 0.30 |
DZANK1 |
double zinc ribbon and ankyrin repeat domains 1 |
427 |
0.47 |
| chr15_71184789_71184970 | 0.29 |
LRRC49 |
leucine rich repeat containing 49 |
86 |
0.56 |
| chr14_101292986_101293251 | 0.29 |
AL117190.2 |
|
2419 |
0.08 |
| chr17_41910293_41910444 | 0.29 |
MPP3 |
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
170 |
0.92 |
| chr14_105525570_105525755 | 0.29 |
GPR132 |
G protein-coupled receptor 132 |
1843 |
0.34 |
| chrX_128888588_128889150 | 0.29 |
XPNPEP2 |
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
15871 |
0.19 |
| chr9_99169894_99170045 | 0.29 |
ZNF367 |
zinc finger protein 367 |
10642 |
0.19 |
| chr12_6601923_6602186 | 0.29 |
NCAPD2 |
non-SMC condensin I complex, subunit D2 |
491 |
0.47 |
| chr15_101099433_101099584 | 0.28 |
RP11-526I2.1 |
|
612 |
0.58 |
| chr16_81870803_81871145 | 0.28 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
45900 |
0.18 |
| chr2_48462400_48462551 | 0.28 |
ENSG00000201010 |
. |
17761 |
0.25 |
| chr19_7929792_7929966 | 0.28 |
EVI5L |
ecotropic viral integration site 5-like |
4408 |
0.09 |
| chr17_75454716_75454867 | 0.28 |
SEPT9 |
septin 9 |
2343 |
0.24 |
| chr21_32519368_32519782 | 0.28 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
17036 |
0.27 |
| chr6_35275141_35275407 | 0.28 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
2241 |
0.29 |
| chr9_35618104_35618377 | 0.27 |
CD72 |
CD72 molecule |
127 |
0.91 |
| chr2_230787566_230787813 | 0.27 |
TRIP12 |
thyroid hormone receptor interactor 12 |
266 |
0.66 |
| chr11_76494030_76494181 | 0.27 |
RP11-21L23.3 |
|
1 |
0.69 |
| chr11_44117733_44117933 | 0.27 |
EXT2 |
exostosin glycosyltransferase 2 |
60 |
0.97 |
| chr3_36985785_36985936 | 0.27 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
688 |
0.69 |
| chr11_15230197_15230348 | 0.27 |
INSC |
inscuteable homolog (Drosophila) |
59618 |
0.15 |
| chr15_29135138_29135296 | 0.26 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
3908 |
0.26 |
| chr1_24117965_24118246 | 0.26 |
LYPLA2 |
lysophospholipase II |
377 |
0.76 |
| chr1_154983991_154984142 | 0.26 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
2858 |
0.1 |
| chr7_150757151_150757478 | 0.26 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
157 |
0.88 |
| chrX_47488098_47488605 | 0.26 |
CFP |
complement factor properdin |
943 |
0.43 |
| chr12_4392748_4393028 | 0.26 |
CCND2-AS1 |
CCND2 antisense RNA 1 |
7538 |
0.17 |
| chr7_127372868_127373019 | 0.26 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
80709 |
0.09 |
| chr19_31841356_31841519 | 0.26 |
TSHZ3 |
teashirt zinc finger homeobox 3 |
984 |
0.54 |
| chr19_16768091_16768274 | 0.26 |
CTC-429P9.4 |
Small integral membrane protein 7; Uncharacterized protein |
2733 |
0.13 |
| chr19_19316868_19317019 | 0.26 |
NR2C2AP |
nuclear receptor 2C2-associated protein |
2710 |
0.13 |
| chr2_63286662_63286820 | 0.26 |
OTX1 |
orthodenticle homeobox 1 |
8804 |
0.23 |
| chr16_1019409_1019579 | 0.26 |
LMF1 |
lipase maturation factor 1 |
1372 |
0.31 |
| chr21_32929864_32930131 | 0.26 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
1293 |
0.39 |
| chr7_150174498_150174649 | 0.26 |
GIMAP8 |
GTPase, IMAP family member 8 |
26855 |
0.14 |
| chr7_151534819_151535015 | 0.26 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
22991 |
0.15 |
| chr5_138851413_138851689 | 0.25 |
AC138517.1 |
Uncharacterized protein |
557 |
0.68 |
| chr12_57635535_57635686 | 0.25 |
NDUFA4L2 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
1135 |
0.31 |
| chr2_71356997_71357205 | 0.25 |
MCEE |
methylmalonyl CoA epimerase |
243 |
0.59 |
| chr1_38454975_38455189 | 0.25 |
SF3A3 |
splicing factor 3a, subunit 3, 60kDa |
577 |
0.62 |
| chr14_98441802_98442041 | 0.25 |
C14orf64 |
chromosome 14 open reading frame 64 |
2462 |
0.45 |
| chr17_19015396_19015547 | 0.25 |
ENSG00000262202 |
. |
478 |
0.65 |
| chr5_142149375_142149625 | 0.25 |
ARHGAP26 |
Rho GTPase activating protein 26 |
449 |
0.88 |
| chr11_74169959_74170165 | 0.25 |
KCNE3 |
potassium voltage-gated channel, Isk-related family, member 3 |
3350 |
0.16 |
| chr5_126317848_126317999 | 0.25 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
48577 |
0.17 |
| chr3_53202293_53202538 | 0.25 |
PRKCD |
protein kinase C, delta |
3270 |
0.22 |
| chr11_117186964_117187239 | 0.25 |
BACE1 |
beta-site APP-cleaving enzyme 1 |
126 |
0.94 |
| chr2_87769897_87770221 | 0.24 |
RP11-1399P15.1 |
|
7494 |
0.31 |
| chr20_55974994_55975197 | 0.24 |
RP4-800J21.3 |
|
6977 |
0.16 |
| chr3_52255710_52256010 | 0.24 |
TLR9 |
toll-like receptor 9 |
4319 |
0.11 |
| chr1_145395576_145396119 | 0.24 |
ENSG00000201558 |
. |
13090 |
0.14 |
| chr11_1801564_1801715 | 0.24 |
CTSD |
cathepsin D |
16417 |
0.08 |
| chr18_20716662_20716943 | 0.24 |
ENSG00000222999 |
. |
326 |
0.57 |
| chr9_95783100_95783251 | 0.24 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
5833 |
0.21 |
| chr14_91814952_91815599 | 0.24 |
ENSG00000265856 |
. |
15218 |
0.22 |
| chr13_24829233_24829410 | 0.24 |
SPATA13-AS1 |
SPATA13 antisense RNA 1 |
744 |
0.63 |
| chr16_89927127_89927278 | 0.24 |
TCF25 |
transcription factor 25 (basic helix-loop-helix) |
12812 |
0.13 |
| chr15_68375595_68375936 | 0.24 |
PIAS1 |
protein inhibitor of activated STAT, 1 |
28851 |
0.22 |
| chr10_63870514_63870665 | 0.24 |
ENSG00000221094 |
. |
5385 |
0.29 |
| chr12_66219604_66219755 | 0.24 |
HMGA2 |
high mobility group AT-hook 2 |
776 |
0.69 |
| chr10_17272231_17272382 | 0.24 |
VIM |
vimentin |
302 |
0.5 |
| chrX_106448945_106449104 | 0.24 |
NUP62CL |
nucleoporin 62kDa C-terminal like |
639 |
0.5 |
| chr20_61638770_61638921 | 0.24 |
BHLHE23 |
basic helix-loop-helix family, member e23 |
458 |
0.81 |
| chr3_183979642_183979943 | 0.24 |
CAMK2N2 |
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
541 |
0.6 |
| chr14_21150005_21150238 | 0.23 |
RNASE4 |
ribonuclease, RNase A family, 4 |
2138 |
0.14 |
| chr9_99983071_99983222 | 0.23 |
CCDC180 |
coiled-coil domain containing 180 |
17619 |
0.21 |
| chr7_33701123_33701274 | 0.23 |
RP11-89N17.1 |
HCG1643653; Uncharacterized protein |
64395 |
0.13 |
| chr20_32837057_32837353 | 0.23 |
ASIP |
agouti signaling protein |
10966 |
0.17 |
| chr7_27205594_27205875 | 0.23 |
HOXA9 |
homeobox A9 |
585 |
0.43 |
| chr9_132816429_132816580 | 0.23 |
GPR107 |
G protein-coupled receptor 107 |
519 |
0.79 |
| chr3_138067657_138067808 | 0.23 |
MRAS |
muscle RAS oncogene homolog |
40 |
0.98 |
| chr12_6876238_6876445 | 0.23 |
MLF2 |
myeloid leukemia factor 2 |
300 |
0.67 |
| chr17_74269472_74269623 | 0.23 |
QRICH2 |
glutamine rich 2 |
6891 |
0.13 |
| chr5_114545827_114545978 | 0.23 |
CTC-428G20.2 |
|
21642 |
0.16 |
| chr9_14315059_14315375 | 0.23 |
NFIB |
nuclear factor I/B |
636 |
0.73 |
| chr5_14596125_14596582 | 0.23 |
FAM105A |
family with sequence similarity 105, member A |
14469 |
0.24 |
| chr7_150674322_150674753 | 0.23 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
477 |
0.73 |
| chr17_77777762_77777981 | 0.22 |
CBX8 |
chromobox homolog 8 |
2539 |
0.19 |
| chr8_146125496_146125701 | 0.22 |
ZNF250 |
zinc finger protein 250 |
1209 |
0.41 |
| chr2_171676330_171676481 | 0.22 |
AC007405.8 |
|
1858 |
0.27 |
| chr7_21985572_21985928 | 0.22 |
CDCA7L |
cell division cycle associated 7-like |
48 |
0.99 |
| chr9_131669773_131669924 | 0.22 |
PHYHD1 |
phytanoyl-CoA dioxygenase domain containing 1 |
13326 |
0.11 |
| chr22_46068446_46068662 | 0.22 |
ATXN10 |
ataxin 10 |
875 |
0.64 |
| chr8_145002100_145002251 | 0.22 |
PLEC |
plectin |
996 |
0.39 |
| chr19_52996325_52996476 | 0.22 |
ZNF578 |
zinc finger protein 578 |
317 |
0.84 |
| chr2_29236792_29237319 | 0.22 |
FAM179A |
family with sequence similarity 179, member A |
4072 |
0.19 |
| chr4_126236035_126236377 | 0.21 |
FAT4 |
FAT atypical cadherin 4 |
1348 |
0.58 |
| chr9_129387391_129387562 | 0.21 |
LMX1B |
LIM homeobox transcription factor 1, beta |
10728 |
0.16 |
| chr22_37623875_37624507 | 0.21 |
SSTR3 |
somatostatin receptor 3 |
15829 |
0.12 |
| chr17_48134073_48134477 | 0.21 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
471 |
0.75 |
| chr3_157155814_157155965 | 0.21 |
PTX3 |
pentraxin 3, long |
1311 |
0.55 |
| chr19_45908936_45909178 | 0.21 |
CD3EAP |
CD3e molecule, epsilon associated protein |
410 |
0.51 |
| chr1_6055265_6055416 | 0.21 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
2569 |
0.24 |
| chr3_193987519_193987707 | 0.21 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
84434 |
0.08 |
| chr18_74811464_74812005 | 0.21 |
MBP |
myelin basic protein |
5483 |
0.32 |
| chr11_118811293_118811572 | 0.21 |
ENSG00000239726 |
. |
9512 |
0.08 |
| chrX_150153284_150153533 | 0.21 |
HMGB3 |
high mobility group box 3 |
596 |
0.75 |
| chr17_75889796_75889947 | 0.21 |
FLJ45079 |
|
11212 |
0.22 |
| chrX_135230186_135230374 | 0.21 |
FHL1 |
four and a half LIM domains 1 |
10 |
0.98 |
| chr19_50063674_50064142 | 0.21 |
NOSIP |
nitric oxide synthase interacting protein |
21 |
0.93 |
| chr9_100617168_100617452 | 0.21 |
FOXE1 |
forkhead box E1 (thyroid transcription factor 2) |
1774 |
0.38 |
| chr14_24837393_24837740 | 0.21 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
19 |
0.94 |
| chr2_106804645_106804858 | 0.21 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
6004 |
0.25 |
| chr1_21978624_21978775 | 0.20 |
RAP1GAP |
RAP1 GTPase activating protein |
351 |
0.89 |
| chr2_231437706_231437857 | 0.20 |
AC010149.4 |
|
6867 |
0.2 |
| chr7_1456845_1457076 | 0.20 |
MICALL2 |
MICAL-like 2 |
42002 |
0.12 |
| chr19_1450029_1450180 | 0.20 |
APC2 |
adenomatosis polyposis coli 2 |
44 |
0.93 |
| chr16_57176887_57177289 | 0.20 |
CPNE2 |
copine II |
23977 |
0.12 |
| chr20_19997607_19997892 | 0.20 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
11 |
0.98 |
| chr3_45187270_45187501 | 0.20 |
CDCP1 |
CUB domain containing protein 1 |
529 |
0.82 |
| chr11_36616334_36616601 | 0.20 |
C11orf74 |
chromosome 11 open reading frame 74 |
67 |
0.63 |
| chr10_63663919_63664225 | 0.20 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
3013 |
0.33 |
| chr1_150550855_150551146 | 0.20 |
MCL1 |
myeloid cell leukemia sequence 1 (BCL2-related) |
1006 |
0.32 |
| chr12_57636992_57637222 | 0.20 |
NDUFA4L2 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
2632 |
0.14 |
| chr19_56014839_56015099 | 0.20 |
SSC5D |
scavenger receptor cysteine rich domain containing (5 domains) |
9711 |
0.08 |
| chr12_96184490_96184641 | 0.20 |
NTN4 |
netrin 4 |
23 |
0.97 |
| chr18_45693523_45693802 | 0.20 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
1725 |
0.38 |
| chr20_47379698_47379939 | 0.20 |
ENSG00000251876 |
. |
23833 |
0.24 |
| chr17_55360579_55360787 | 0.20 |
MSI2 |
musashi RNA-binding protein 2 |
2522 |
0.39 |
| chr2_6924565_6924716 | 0.20 |
AC017076.5 |
|
49022 |
0.15 |
| chr5_1799046_1799396 | 0.20 |
MRPL36 |
mitochondrial ribosomal protein L36 |
643 |
0.67 |
| chr17_46805969_46806120 | 0.19 |
HOXB13 |
homeobox B13 |
496 |
0.65 |
| chr8_8670148_8670299 | 0.19 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
80932 |
0.08 |
| chr2_61272587_61272742 | 0.19 |
KIAA1841 |
KIAA1841 |
20404 |
0.15 |
| chr2_240182188_240182405 | 0.19 |
ENSG00000265215 |
. |
44861 |
0.12 |
| chr10_7860991_7861192 | 0.19 |
TAF3 |
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa |
624 |
0.76 |
| chr2_62425937_62426123 | 0.19 |
B3GNT2 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
2782 |
0.24 |
| chr18_60382474_60382625 | 0.19 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
123 |
0.97 |
| chr9_22446679_22446830 | 0.19 |
DMRTA1 |
DMRT-like family A1 |
86 |
0.99 |
| chr2_63284454_63284605 | 0.19 |
OTX1 |
orthodenticle homeobox 1 |
6592 |
0.24 |
| chr13_99703300_99703553 | 0.19 |
ENSG00000207298 |
. |
25938 |
0.17 |
| chr20_30639419_30639649 | 0.19 |
HCK |
hemopoietic cell kinase |
457 |
0.65 |
| chr5_1799598_1799749 | 0.19 |
MRPL36 |
mitochondrial ribosomal protein L36 |
191 |
0.93 |
| chr16_88766157_88766414 | 0.19 |
RNF166 |
ring finger protein 166 |
171 |
0.86 |
| chr3_32479509_32479795 | 0.19 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
46121 |
0.14 |
| chr6_153303203_153303466 | 0.19 |
FBXO5 |
F-box protein 5 |
819 |
0.52 |
| chr15_92404124_92404311 | 0.19 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
6866 |
0.27 |
| chrX_55027132_55027456 | 0.19 |
APEX2 |
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
504 |
0.73 |
| chr17_30335102_30335253 | 0.19 |
LRRC37B |
leucine rich repeat containing 37B |
170 |
0.95 |
| chr17_76110986_76111137 | 0.19 |
ENSG00000204282 |
. |
3181 |
0.16 |
| chr2_7061705_7061856 | 0.19 |
ENSG00000228203 |
. |
2940 |
0.24 |
| chr19_36443027_36443178 | 0.19 |
LRFN3 |
leucine rich repeat and fibronectin type III domain containing 3 |
15080 |
0.09 |
| chr1_28586799_28587133 | 0.19 |
SESN2 |
sestrin 2 |
928 |
0.45 |
| chr12_64061940_64062091 | 0.19 |
DPY19L2 |
dpy-19-like 2 (C. elegans) |
300 |
0.93 |
| chr20_60962587_60962934 | 0.19 |
RPS21 |
ribosomal protein S21 |
379 |
0.79 |
| chr2_110371414_110371565 | 0.19 |
SEPT10 |
septin 10 |
3 |
0.88 |
| chr22_30663660_30663811 | 0.19 |
OSM |
oncostatin M |
906 |
0.4 |
| chr15_91138025_91138489 | 0.19 |
CRTC3 |
CREB regulated transcription coactivator 3 |
1312 |
0.32 |
| chr1_145610669_145610820 | 0.19 |
POLR3C |
polymerase (RNA) III (DNA directed) polypeptide C (62kD) |
140 |
0.69 |
| chr3_153839356_153839507 | 0.19 |
ARHGEF26 |
Rho guanine nucleotide exchange factor (GEF) 26 |
202 |
0.64 |
| chr9_139948636_139948800 | 0.19 |
ENTPD2 |
ectonucleoside triphosphate diphosphohydrolase 2 |
221 |
0.75 |
| chr14_89830784_89831443 | 0.19 |
RP11-356K23.2 |
|
9709 |
0.17 |
| chr3_9177794_9177945 | 0.19 |
SRGAP3-AS2 |
SRGAP3 antisense RNA 2 |
56308 |
0.12 |
| chr15_31622016_31622167 | 0.19 |
KLF13 |
Kruppel-like factor 13 |
3033 |
0.39 |
| chr19_8386904_8387224 | 0.19 |
RPS28 |
ribosomal protein S28 |
680 |
0.38 |
| chr3_124449486_124449637 | 0.19 |
UMPS |
uridine monophosphate synthetase |
262 |
0.87 |
| chr10_73514684_73515259 | 0.19 |
C10orf54 |
chromosome 10 open reading frame 54 |
2416 |
0.29 |
| chr16_3640725_3640876 | 0.18 |
NLRC3 |
NLR family, CARD domain containing 3 |
13399 |
0.13 |
| chr17_45266002_45266457 | 0.18 |
CDC27 |
cell division cycle 27 |
313 |
0.84 |
| chr16_3122337_3122494 | 0.18 |
ENSG00000252561 |
. |
2891 |
0.09 |
| chrX_39712527_39712734 | 0.18 |
ENSG00000263972 |
. |
15815 |
0.27 |
| chr1_209957444_209957681 | 0.18 |
C1orf74 |
chromosome 1 open reading frame 74 |
342 |
0.82 |
| chr20_3062971_3063207 | 0.18 |
AVP |
arginine vasopressin |
2281 |
0.19 |
| chr13_40177019_40177302 | 0.18 |
LHFP |
lipoma HMGIC fusion partner |
148 |
0.97 |
| chr11_75294844_75295036 | 0.18 |
CTD-2530H12.4 |
|
699 |
0.6 |
| chr2_68546906_68547171 | 0.18 |
CNRIP1 |
cannabinoid receptor interacting protein 1 |
19 |
0.97 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.2 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.1 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.2 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.2 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.1 | 0.2 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.2 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.1 | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process(GO:0009130) |
| 0.1 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 0.2 | GO:0021778 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.3 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.4 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.2 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.2 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.2 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.2 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.2 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.1 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0052033 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.0 | GO:0060510 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0061140 | lung secretory cell differentiation(GO:0061140) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0085029 | extracellular matrix assembly(GO:0085029) |
| 0.0 | 0.0 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.2 | GO:0042772 | DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.2 | GO:0090114 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.0 | GO:0035872 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.2 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.0 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.3 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0032621 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.2 | GO:0009067 | aspartate family amino acid biosynthetic process(GO:0009067) |
| 0.0 | 0.1 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0035930 | obsolete regulation of natriuresis(GO:0003078) corticosteroid hormone secretion(GO:0035930) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) |
| 0.0 | 0.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.0 | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway(GO:0031664) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0042511 | positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 | 0.0 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.0 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.0 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0060391 | regulation of SMAD protein import into nucleus(GO:0060390) positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0060678 | dichotomous subdivision of terminal units involved in ureteric bud branching(GO:0060678) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.0 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.0 | 0.0 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.1 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.0 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.0 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 0.0 | GO:0051153 | regulation of striated muscle cell differentiation(GO:0051153) |
| 0.0 | 0.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.1 | GO:0006862 | nucleotide transport(GO:0006862) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.0 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.1 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.0 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.0 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.0 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.3 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.0 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.0 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0001228 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.0 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |