Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
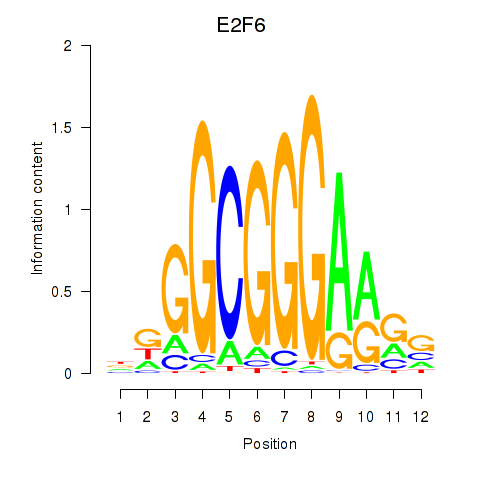
Results for E2F6
Z-value: 1.11
Transcription factors associated with E2F6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F6
|
ENSG00000169016.12 | E2F transcription factor 6 |
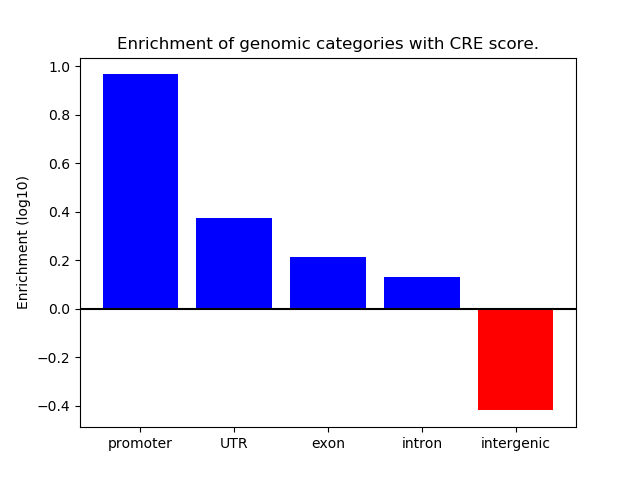
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_11623681_11623832 | E2F6 | 17481 | 0.131240 | -0.52 | 1.5e-01 | Click! |
| chr2_11622106_11622257 | E2F6 | 15906 | 0.134813 | -0.51 | 1.6e-01 | Click! |
| chr2_11605347_11605717 | E2F6 | 473 | 0.729351 | -0.48 | 1.9e-01 | Click! |
| chr2_11622351_11622584 | E2F6 | 16192 | 0.134166 | -0.40 | 2.9e-01 | Click! |
| chr2_11622769_11622930 | E2F6 | 16574 | 0.133300 | -0.39 | 3.0e-01 | Click! |
Activity of the E2F6 motif across conditions
Conditions sorted by the z-value of the E2F6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
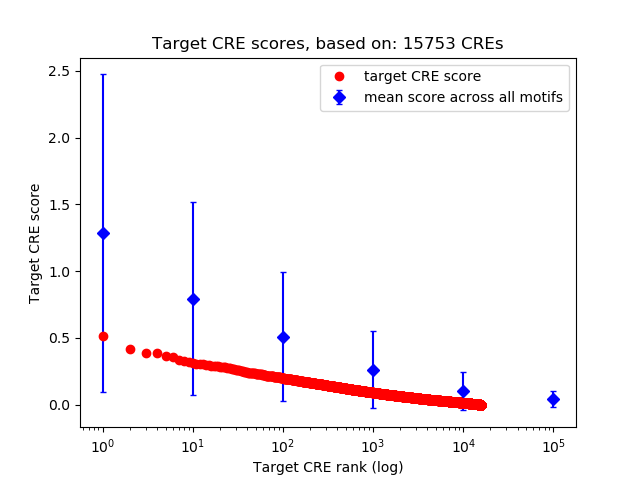
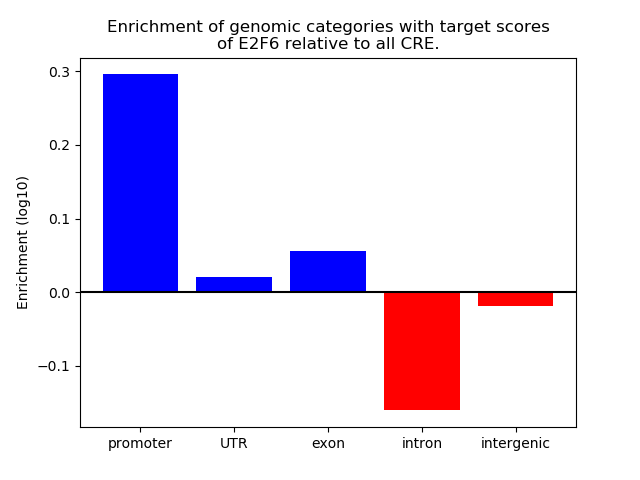
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_39179164_39179315 | 0.52 |
SUN2 |
Sad1 and UNC84 domain containing 2 |
10909 |
0.09 |
| chr1_8931641_8932436 | 0.42 |
ENO1-IT1 |
ENO1 intronic transcript 1 (non-protein coding) |
6028 |
0.14 |
| chr17_75311699_75312099 | 0.39 |
SEPT9 |
septin 9 |
3698 |
0.27 |
| chr1_1476104_1476623 | 0.39 |
TMEM240 |
transmembrane protein 240 |
530 |
0.63 |
| chr2_112896432_112896734 | 0.36 |
FBLN7 |
fibulin 7 |
423 |
0.88 |
| chr17_36862672_36863533 | 0.35 |
CTB-58E17.3 |
|
588 |
0.44 |
| chr9_128507884_128508777 | 0.33 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
1294 |
0.56 |
| chr12_6657819_6658220 | 0.33 |
IFFO1 |
intermediate filament family orphan 1 |
3047 |
0.1 |
| chr21_35184316_35184478 | 0.32 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
12170 |
0.23 |
| chr1_228603985_228604375 | 0.31 |
TRIM17 |
tripartite motif containing 17 |
148 |
0.9 |
| chr17_77788487_77789261 | 0.31 |
ENSG00000238331 |
. |
4396 |
0.16 |
| chr19_850886_851037 | 0.31 |
ELANE |
elastase, neutrophil expressed |
53 |
0.94 |
| chr1_156675699_156676565 | 0.31 |
CRABP2 |
cellular retinoic acid binding protein 2 |
524 |
0.6 |
| chr3_53272501_53272842 | 0.30 |
TKT |
transketolase |
17345 |
0.13 |
| chr6_81164529_81164680 | 0.30 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
348221 |
0.01 |
| chr15_83378361_83379204 | 0.29 |
AP3B2 |
adaptor-related protein complex 3, beta 2 subunit |
116 |
0.95 |
| chr12_132315122_132315695 | 0.29 |
MMP17 |
matrix metallopeptidase 17 (membrane-inserted) |
2323 |
0.26 |
| chr17_55945832_55946020 | 0.29 |
CUEDC1 |
CUE domain containing 1 |
1150 |
0.47 |
| chr1_54686195_54686878 | 0.29 |
MRPL37 |
mitochondrial ribosomal protein L37 |
10917 |
0.13 |
| chr8_99170182_99170517 | 0.29 |
ENSG00000252558 |
. |
34192 |
0.13 |
| chr1_44031055_44031511 | 0.28 |
PTPRF |
protein tyrosine phosphatase, receptor type, F |
20536 |
0.18 |
| chr10_88160229_88160748 | 0.28 |
GRID1 |
glutamate receptor, ionotropic, delta 1 |
34253 |
0.17 |
| chr17_56356237_56356388 | 0.28 |
MPO |
myeloperoxidase |
1984 |
0.23 |
| chr15_90563466_90563923 | 0.28 |
ENSG00000265871 |
. |
13707 |
0.14 |
| chr10_123702458_123702803 | 0.28 |
ATE1 |
arginyltransferase 1 |
14314 |
0.17 |
| chr11_69066370_69066772 | 0.27 |
MYEOV |
myeloma overexpressed |
4946 |
0.32 |
| chr17_38082798_38082949 | 0.27 |
ORMDL3 |
ORM1-like 3 (S. cerevisiae) |
221 |
0.89 |
| chr1_203122833_203123125 | 0.27 |
RP11-335O13.8 |
|
951 |
0.45 |
| chr1_68237207_68237358 | 0.26 |
ENSG00000238778 |
. |
1054 |
0.59 |
| chr13_53313127_53313462 | 0.26 |
LECT1 |
leukocyte cell derived chemotaxin 1 |
193 |
0.96 |
| chr19_7989733_7990535 | 0.26 |
CTD-3193O13.8 |
|
752 |
0.29 |
| chr5_76925321_76925875 | 0.26 |
WDR41 |
WD repeat domain 41 |
9162 |
0.23 |
| chr4_157594425_157594576 | 0.26 |
RP11-171N4.3 |
|
30768 |
0.2 |
| chr22_41843929_41844303 | 0.26 |
TOB2 |
transducer of ERBB2, 2 |
1089 |
0.43 |
| chr9_36165368_36165519 | 0.25 |
CCIN |
calicin |
3946 |
0.21 |
| chr9_139519589_139519822 | 0.25 |
ENSG00000252440 |
. |
22738 |
0.09 |
| chr14_71276457_71276666 | 0.25 |
MAP3K9 |
mitogen-activated protein kinase kinase kinase 9 |
310 |
0.94 |
| chr2_87016233_87016699 | 0.25 |
CD8A |
CD8a molecule |
1482 |
0.41 |
| chr10_105452470_105452921 | 0.24 |
SH3PXD2A |
SH3 and PX domains 2A |
235 |
0.93 |
| chr11_3175380_3175656 | 0.24 |
OSBPL5 |
oxysterol binding protein-like 5 |
7105 |
0.16 |
| chr5_180480179_180480493 | 0.24 |
BTNL9 |
butyrophilin-like 9 |
13070 |
0.16 |
| chr11_12836667_12836952 | 0.24 |
RP11-47J17.3 |
|
8405 |
0.22 |
| chr22_45521390_45521628 | 0.24 |
NUP50 |
nucleoporin 50kDa |
38213 |
0.14 |
| chr5_73556832_73556983 | 0.24 |
ENSG00000222551 |
. |
10683 |
0.28 |
| chr22_50422080_50422460 | 0.24 |
IL17REL |
interleukin 17 receptor E-like |
24851 |
0.15 |
| chr22_36828593_36829238 | 0.24 |
ENSG00000252575 |
. |
6056 |
0.16 |
| chr5_172327841_172328220 | 0.24 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
4190 |
0.22 |
| chr10_81027570_81027806 | 0.23 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
38287 |
0.19 |
| chr4_77675626_77675892 | 0.23 |
RP11-359D14.2 |
|
3868 |
0.27 |
| chr9_139510576_139510771 | 0.23 |
ENSG00000252440 |
. |
13706 |
0.1 |
| chr20_58520957_58521211 | 0.23 |
FAM217B |
family with sequence similarity 217, member B |
3905 |
0.19 |
| chr19_1513130_1513873 | 0.23 |
ADAMTSL5 |
ADAMTS-like 5 |
313 |
0.73 |
| chr6_11044071_11044432 | 0.23 |
ELOVL2-AS1 |
ELOVL2 antisense RNA 1 |
258 |
0.53 |
| chr17_32574614_32574944 | 0.23 |
CCL2 |
chemokine (C-C motif) ligand 2 |
7525 |
0.13 |
| chr11_925206_925651 | 0.23 |
AP2A2 |
adaptor-related protein complex 2, alpha 2 subunit |
444 |
0.71 |
| chr3_120180997_120181426 | 0.23 |
FSTL1 |
follistatin-like 1 |
11111 |
0.26 |
| chr12_65700883_65701407 | 0.23 |
MSRB3 |
methionine sulfoxide reductase B3 |
19510 |
0.22 |
| chr1_114525815_114526112 | 0.23 |
OLFML3 |
olfactomedin-like 3 |
3268 |
0.23 |
| chr8_105478631_105479267 | 0.23 |
DPYS |
dihydropyrimidinase |
323 |
0.87 |
| chr2_25475353_25475794 | 0.23 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
393 |
0.88 |
| chr19_728237_728510 | 0.22 |
PALM |
paralemmin |
19272 |
0.09 |
| chr6_154640196_154640597 | 0.22 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
10819 |
0.3 |
| chr2_174053187_174053569 | 0.22 |
MLK7-AS1 |
MLK7 antisense RNA 1 |
18962 |
0.26 |
| chr2_20715345_20715496 | 0.22 |
ENSG00000200829 |
. |
34815 |
0.16 |
| chr22_45622655_45622806 | 0.22 |
KIAA0930 |
KIAA0930 |
54 |
0.97 |
| chr17_48114815_48114966 | 0.22 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
18449 |
0.13 |
| chr9_124581839_124582120 | 0.22 |
RP11-244O19.1 |
|
248 |
0.95 |
| chr21_47485945_47486454 | 0.22 |
AP001471.1 |
|
31245 |
0.13 |
| chr9_132096828_132097338 | 0.22 |
ENSG00000242281 |
. |
35657 |
0.16 |
| chr12_120662468_120662643 | 0.21 |
PXN |
paxillin |
2 |
0.96 |
| chr16_24679897_24680048 | 0.21 |
TNRC6A |
trinucleotide repeat containing 6A |
61044 |
0.14 |
| chr10_126791455_126791637 | 0.21 |
CTBP2 |
C-terminal binding protein 2 |
55739 |
0.14 |
| chr17_80655614_80655903 | 0.21 |
RAB40B |
RAB40B, member RAS oncogene family |
780 |
0.49 |
| chr12_63544901_63545298 | 0.21 |
AVPR1A |
arginine vasopressin receptor 1A |
377 |
0.93 |
| chr1_8676963_8677114 | 0.21 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
7401 |
0.28 |
| chr9_14322498_14322883 | 0.21 |
NFIB |
nuclear factor I/B |
353 |
0.91 |
| chr17_72039340_72039491 | 0.21 |
RPL38 |
ribosomal protein L38 |
160306 |
0.03 |
| chr17_37353315_37353878 | 0.21 |
CACNB1 |
calcium channel, voltage-dependent, beta 1 subunit |
359 |
0.79 |
| chr3_23653779_23653992 | 0.21 |
ENSG00000252562 |
. |
1365 |
0.51 |
| chr9_140009377_140009643 | 0.21 |
DPP7 |
dipeptidyl-peptidase 7 |
119 |
0.88 |
| chr3_120279483_120279766 | 0.21 |
NDUFB4 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
35532 |
0.19 |
| chr3_190580663_190581079 | 0.21 |
GMNC |
geminin coiled-coil domain containing |
467 |
0.9 |
| chr3_42692749_42693153 | 0.21 |
ZBTB47 |
zinc finger and BTB domain containing 47 |
2225 |
0.17 |
| chr4_157996843_157997211 | 0.21 |
GLRB |
glycine receptor, beta |
182 |
0.96 |
| chr4_4388363_4388660 | 0.21 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
246 |
0.94 |
| chr10_6243827_6244661 | 0.21 |
RP11-414H17.5 |
|
412 |
0.58 |
| chr2_86450156_86450475 | 0.21 |
MRPL35 |
mitochondrial ribosomal protein L35 |
23729 |
0.12 |
| chr9_137118247_137118684 | 0.21 |
ENSG00000221676 |
. |
88779 |
0.07 |
| chr7_127306838_127306989 | 0.21 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
14679 |
0.17 |
| chr3_120278487_120278806 | 0.21 |
NDUFB4 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
36510 |
0.19 |
| chr4_20432301_20432452 | 0.20 |
SLIT2-IT1 |
SLIT2 intronic transcript 1 (non-protein coding) |
38564 |
0.21 |
| chr6_44044467_44044818 | 0.20 |
RP5-1120P11.1 |
|
2253 |
0.28 |
| chr12_26428456_26429005 | 0.20 |
RP11-283G6.5 |
|
3877 |
0.25 |
| chr2_129062963_129063193 | 0.20 |
HS6ST1 |
heparan sulfate 6-O-sulfotransferase 1 |
13073 |
0.24 |
| chr17_41016311_41016609 | 0.20 |
AOC4P |
amine oxidase, copper containing 4, pseudogene |
2836 |
0.12 |
| chr17_42872109_42872587 | 0.20 |
GJC1 |
gap junction protein, gamma 1, 45kDa |
9986 |
0.14 |
| chr1_15273356_15273566 | 0.20 |
KAZN |
kazrin, periplakin interacting protein |
1046 |
0.69 |
| chr1_33800790_33801246 | 0.20 |
ENSG00000222112 |
. |
1447 |
0.27 |
| chr7_128531158_128531719 | 0.20 |
ENSG00000221401 |
. |
15387 |
0.13 |
| chr1_19977045_19977345 | 0.20 |
NBL1 |
neuroblastoma 1, DAN family BMP antagonist |
3091 |
0.21 |
| chr16_85932462_85932735 | 0.20 |
IRF8 |
interferon regulatory factor 8 |
171 |
0.96 |
| chr10_81058240_81058391 | 0.20 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
7660 |
0.25 |
| chr4_38566945_38567096 | 0.20 |
RP11-617D20.1 |
|
59176 |
0.13 |
| chr9_84305434_84305645 | 0.20 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
1319 |
0.53 |
| chr1_56942959_56943169 | 0.20 |
ENSG00000223307 |
. |
99974 |
0.08 |
| chr1_236105296_236105530 | 0.19 |
ENSG00000206803 |
. |
26698 |
0.17 |
| chr11_43942650_43942801 | 0.19 |
RP11-613D13.4 |
|
99 |
0.68 |
| chr10_75685865_75686026 | 0.19 |
C10orf55 |
chromosome 10 open reading frame 55 |
3410 |
0.19 |
| chr12_121890704_121890855 | 0.19 |
RNF34 |
ring finger protein 34, E3 ubiquitin protein ligase |
43501 |
0.13 |
| chr1_156120275_156120768 | 0.19 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
711 |
0.53 |
| chr10_102279085_102279348 | 0.19 |
SEC31B |
SEC31 homolog B (S. cerevisiae) |
375 |
0.84 |
| chr16_86806513_86806664 | 0.19 |
FOXL1 |
forkhead box L1 |
194473 |
0.03 |
| chr21_47714776_47715544 | 0.19 |
YBEY |
ybeY metallopeptidase (putative) |
8601 |
0.11 |
| chr8_134388401_134388552 | 0.19 |
NDRG1 |
N-myc downstream regulated 1 |
74211 |
0.11 |
| chr18_76740387_76741175 | 0.19 |
SALL3 |
spalt-like transcription factor 3 |
506 |
0.87 |
| chr20_25604864_25605138 | 0.19 |
ZNF337-AS1 |
ZNF337 antisense RNA 1 |
132 |
0.58 |
| chr16_73104328_73104761 | 0.19 |
ZFHX3 |
zinc finger homeobox 3 |
10947 |
0.24 |
| chr3_171596267_171596418 | 0.19 |
TMEM212-IT1 |
TMEM212 intronic transcript 1 (non-protein coding) |
15884 |
0.2 |
| chr7_107254455_107254606 | 0.19 |
ENSG00000238832 |
. |
10722 |
0.14 |
| chr19_45248669_45248985 | 0.19 |
BCL3 |
B-cell CLL/lymphoma 3 |
2977 |
0.15 |
| chr6_43097250_43097447 | 0.19 |
PTK7 |
protein tyrosine kinase 7 |
981 |
0.43 |
| chr4_13549043_13549422 | 0.19 |
NKX3-2 |
NK3 homeobox 2 |
2558 |
0.32 |
| chr12_125052866_125053278 | 0.19 |
NCOR2 |
nuclear receptor corepressor 2 |
1062 |
0.68 |
| chr17_73521888_73522295 | 0.19 |
LLGL2 |
lethal giant larvae homolog 2 (Drosophila) |
221 |
0.88 |
| chr8_99468654_99468805 | 0.19 |
ENSG00000207378 |
. |
6549 |
0.21 |
| chr7_126893149_126893382 | 0.19 |
GRM8 |
glutamate receptor, metabotropic 8 |
83 |
0.98 |
| chr15_72522895_72523387 | 0.19 |
PKM |
pyruvate kinase, muscle |
313 |
0.87 |
| chr22_44704706_44704857 | 0.19 |
KIAA1644 |
KIAA1644 |
3950 |
0.31 |
| chr11_66102916_66103067 | 0.19 |
RIN1 |
Ras and Rab interactor 1 |
886 |
0.25 |
| chr1_22236518_22236840 | 0.19 |
HSPG2 |
heparan sulfate proteoglycan 2 |
13876 |
0.16 |
| chr7_128431078_128431544 | 0.19 |
CCDC136 |
coiled-coil domain containing 136 |
153 |
0.93 |
| chr19_33790905_33791151 | 0.19 |
CTD-2540B15.11 |
|
188 |
0.87 |
| chr17_76370784_76371498 | 0.19 |
PGS1 |
phosphatidylglycerophosphate synthase 1 |
3580 |
0.16 |
| chr12_104770624_104770775 | 0.19 |
TXNRD1 |
thioredoxin reductase 1 |
67070 |
0.1 |
| chr6_43740851_43741100 | 0.19 |
VEGFA |
vascular endothelial growth factor A |
1115 |
0.46 |
| chr19_1099011_1099283 | 0.18 |
POLR2E |
polymerase (RNA) II (DNA directed) polypeptide E, 25kDa |
3549 |
0.12 |
| chr10_77718805_77719099 | 0.18 |
ENSG00000215921 |
. |
135416 |
0.05 |
| chr22_50344908_50345281 | 0.18 |
PIM3 |
pim-3 oncogene |
9067 |
0.17 |
| chr4_675635_675881 | 0.18 |
MYL5 |
myosin, light chain 5, regulatory |
3780 |
0.12 |
| chr19_35786776_35787077 | 0.18 |
MAG |
myelin associated glycoprotein |
3862 |
0.11 |
| chr14_68864088_68864387 | 0.18 |
RAD51B |
RAD51 paralog B |
13990 |
0.28 |
| chr6_125425186_125425448 | 0.18 |
TPD52L1 |
tumor protein D52-like 1 |
14878 |
0.26 |
| chr5_172297414_172297971 | 0.18 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
34528 |
0.14 |
| chr15_98509107_98509579 | 0.18 |
ARRDC4 |
arrestin domain containing 4 |
5415 |
0.27 |
| chr1_32178967_32179267 | 0.18 |
COL16A1 |
collagen, type XVI, alpha 1 |
9197 |
0.14 |
| chr3_48590196_48590347 | 0.18 |
PFKFB4 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
3985 |
0.12 |
| chr2_9293102_9293360 | 0.18 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
53663 |
0.16 |
| chr11_2847111_2847315 | 0.18 |
KCNQ1-AS1 |
KCNQ1 antisense RNA 1 |
35585 |
0.12 |
| chr11_75140643_75140996 | 0.18 |
KLHL35 |
kelch-like family member 35 |
417 |
0.77 |
| chr12_48276558_48277156 | 0.18 |
VDR |
vitamin D (1,25- dihydroxyvitamin D3) receptor |
139 |
0.8 |
| chr12_56882110_56882566 | 0.18 |
GLS2 |
glutaminase 2 (liver, mitochondrial) |
157 |
0.93 |
| chr11_67781609_67782080 | 0.18 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
4054 |
0.13 |
| chr5_72672425_72672576 | 0.18 |
FOXD1 |
forkhead box D1 |
71852 |
0.1 |
| chr10_129726535_129726686 | 0.18 |
RP11-4C20.4 |
|
6352 |
0.23 |
| chr13_114563717_114564199 | 0.18 |
GAS6 |
growth arrest-specific 6 |
3082 |
0.32 |
| chr1_110363596_110363760 | 0.18 |
EPS8L3 |
EPS8-like 3 |
57029 |
0.08 |
| chr22_46505362_46505513 | 0.18 |
ENSG00000198986 |
. |
3192 |
0.13 |
| chr8_6519065_6519216 | 0.18 |
CTD-2541M15.1 |
|
12812 |
0.22 |
| chr3_194921339_194921490 | 0.18 |
ENSG00000206600 |
. |
14102 |
0.16 |
| chr19_18548066_18548217 | 0.18 |
ISYNA1 |
inositol-3-phosphate synthase 1 |
22 |
0.95 |
| chr14_77384650_77384809 | 0.18 |
ENSG00000223174 |
. |
28123 |
0.17 |
| chr14_23771801_23772188 | 0.18 |
PPP1R3E |
protein phosphatase 1, regulatory subunit 3E |
63 |
0.92 |
| chr6_71958092_71958555 | 0.18 |
RP11-154D6.1 |
|
36312 |
0.16 |
| chr3_184301748_184302108 | 0.18 |
EPHB3 |
EPH receptor B3 |
22356 |
0.17 |
| chr4_79487146_79487361 | 0.17 |
ANXA3 |
annexin A3 |
12179 |
0.24 |
| chr21_44809582_44809733 | 0.17 |
SIK1 |
salt-inducible kinase 1 |
37351 |
0.2 |
| chr10_114881000_114881218 | 0.17 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
22570 |
0.27 |
| chr22_41405475_41405715 | 0.17 |
ENSG00000222698 |
. |
35906 |
0.11 |
| chr7_91788801_91788952 | 0.17 |
LRRD1 |
leucine-rich repeats and death domain containing 1 |
5714 |
0.15 |
| chr13_52769511_52769985 | 0.17 |
MRPS31P5 |
mitochondrial ribosomal protein S31 pseudogene 5 |
1235 |
0.51 |
| chr2_9400376_9400527 | 0.17 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
53557 |
0.16 |
| chr11_27740496_27740764 | 0.17 |
BDNF |
brain-derived neurotrophic factor |
664 |
0.79 |
| chr7_17730022_17730173 | 0.17 |
SNX13 |
sorting nexin 13 |
249994 |
0.02 |
| chr20_62273575_62273807 | 0.17 |
STMN3 |
stathmin-like 3 |
10428 |
0.1 |
| chr9_32431789_32431940 | 0.17 |
ACO1 |
aconitase 1, soluble |
47246 |
0.14 |
| chr6_158142657_158142808 | 0.17 |
SNX9 |
sorting nexin 9 |
101564 |
0.07 |
| chr3_87841746_87841974 | 0.17 |
ENSG00000200703 |
. |
50061 |
0.18 |
| chr2_38084910_38085061 | 0.17 |
RMDN2 |
regulator of microtubule dynamics 2 |
65345 |
0.13 |
| chr2_241506212_241507153 | 0.17 |
RNPEPL1 |
arginyl aminopeptidase (aminopeptidase B)-like 1 |
1422 |
0.26 |
| chr6_6795563_6795740 | 0.17 |
ENSG00000240936 |
. |
143479 |
0.04 |
| chr9_95873753_95873904 | 0.17 |
C9orf89 |
chromosome 9 open reading frame 89 |
15328 |
0.15 |
| chr1_155035496_155036067 | 0.17 |
EFNA4 |
ephrin-A4 |
426 |
0.41 |
| chr2_118981232_118982329 | 0.17 |
INSIG2 |
insulin induced gene 2 |
135730 |
0.05 |
| chr2_10184378_10184666 | 0.17 |
KLF11 |
Kruppel-like factor 11 |
150 |
0.93 |
| chr19_12912283_12912701 | 0.17 |
PRDX2 |
peroxiredoxin 2 |
109 |
0.89 |
| chr16_84956251_84956402 | 0.17 |
ZDHHC7 |
zinc finger, DHHC-type containing 7 |
59397 |
0.1 |
| chr21_46963567_46964113 | 0.17 |
SLC19A1 |
solute carrier family 19 (folate transporter), member 1 |
474 |
0.84 |
| chr15_40544709_40545241 | 0.17 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
114 |
0.52 |
| chr7_5458267_5458922 | 0.17 |
AC093620.5 |
|
864 |
0.36 |
| chr1_40423957_40424317 | 0.17 |
MFSD2A |
major facilitator superfamily domain containing 2A |
3315 |
0.2 |
| chr6_155053378_155054103 | 0.17 |
SCAF8 |
SR-related CTD-associated factor 8 |
719 |
0.76 |
| chr19_39919469_39919620 | 0.17 |
PLEKHG2 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
4133 |
0.09 |
| chr17_983655_983915 | 0.17 |
ABR |
active BCR-related |
1399 |
0.45 |
| chr15_67316146_67316685 | 0.17 |
SMAD3 |
SMAD family member 3 |
39686 |
0.2 |
| chr16_4323567_4324032 | 0.17 |
TFAP4 |
transcription factor AP-4 (activating enhancer binding protein 4) |
723 |
0.63 |
| chr16_521494_521645 | 0.17 |
RAB11FIP3 |
RAB11 family interacting protein 3 (class II) |
3281 |
0.13 |
| chr20_48920943_48921193 | 0.17 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
113692 |
0.05 |
| chr14_25518403_25519489 | 0.17 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
149 |
0.98 |
| chr20_4230129_4230416 | 0.17 |
ADRA1D |
adrenoceptor alpha 1D |
551 |
0.82 |
| chr5_392750_392913 | 0.17 |
AHRR |
aryl-hydrocarbon receptor repressor |
28204 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.3 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.1 | 0.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.2 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.1 | GO:0014055 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.1 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.1 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.0 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.2 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.3 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 0.2 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.0 | GO:0046084 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0021843 | substrate-independent telencephalic tangential migration(GO:0021826) interneuron migration from the subpallium to the cortex(GO:0021830) substrate-independent telencephalic tangential interneuron migration(GO:0021843) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.2 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.1 | GO:0072160 | nephron tubule epithelial cell differentiation(GO:0072160) regulation of nephron tubule epithelial cell differentiation(GO:0072182) regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.0 | GO:0071605 | monocyte chemotactic protein-1 production(GO:0071605) regulation of monocyte chemotactic protein-1 production(GO:0071637) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:1903670 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0060087 | relaxation of smooth muscle(GO:0044557) negative regulation of smooth muscle contraction(GO:0045986) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0033133 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.2 | GO:0071450 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.0 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0060431 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) primary lung bud formation(GO:0060431) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.0 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.0 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.3 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0002885 | positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.0 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.0 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.1 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.0 | 0.1 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.2 | GO:0051350 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) |
| 0.0 | 0.2 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.2 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.3 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 0.0 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway(GO:0033145) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0031034 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.0 | GO:1901374 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.0 | 0.1 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:1900087 | traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.0 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0006837 | serotonin transport(GO:0006837) |
| 0.0 | 0.0 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.1 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0032352 | positive regulation of hormone metabolic process(GO:0032352) positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.0 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.0 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 1.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0051380 | adrenergic receptor activity(GO:0004935) beta-adrenergic receptor activity(GO:0004939) epinephrine binding(GO:0051379) norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.1 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0004601 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.0 | 0.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.0 | REACTOME OPIOID SIGNALLING | Genes involved in Opioid Signalling |
| 0.0 | 0.1 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |