Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
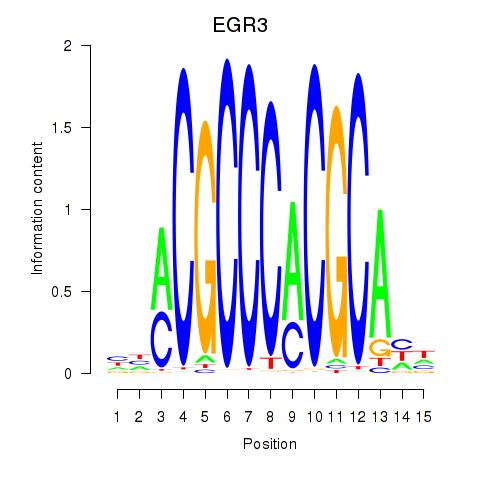
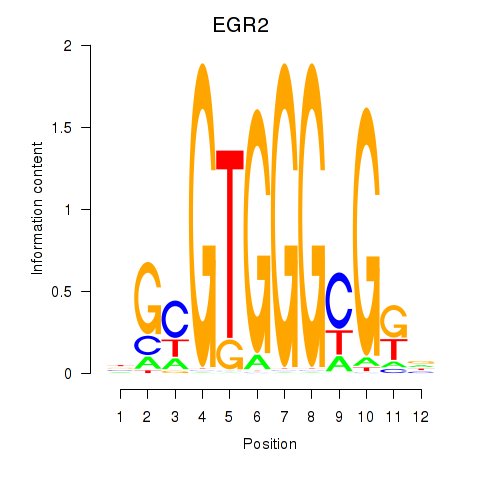
Results for EGR3_EGR2
Z-value: 1.34
Transcription factors associated with EGR3_EGR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR3
|
ENSG00000179388.8 | early growth response 3 |
|
EGR2
|
ENSG00000122877.9 | early growth response 2 |
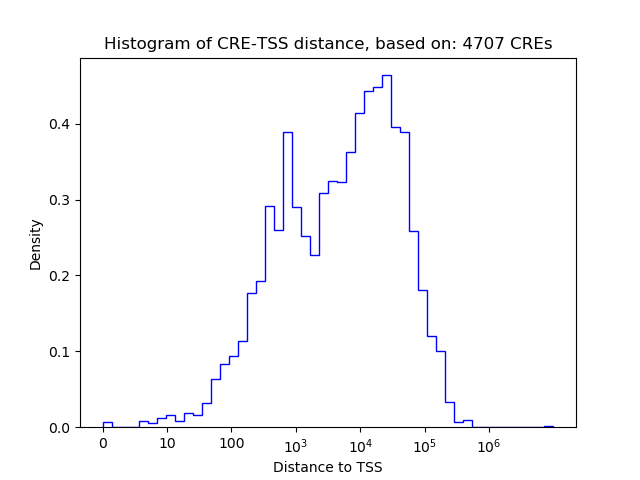
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_64586125_64586276 | EGR2 | 7273 | 0.209296 | -0.74 | 2.2e-02 | Click! |
| chr10_64612517_64612668 | EGR2 | 33665 | 0.171609 | -0.73 | 2.5e-02 | Click! |
| chr10_64577192_64577553 | EGR2 | 1246 | 0.474352 | -0.68 | 4.3e-02 | Click! |
| chr10_64610890_64611041 | EGR2 | 32038 | 0.174430 | -0.65 | 5.7e-02 | Click! |
| chr10_64577644_64577796 | EGR2 | 1207 | 0.486599 | -0.61 | 8.2e-02 | Click! |
| chr8_22561380_22561809 | EGR3 | 10779 | 0.132013 | -0.89 | 1.4e-03 | Click! |
| chr8_22552857_22553008 | EGR3 | 2117 | 0.218138 | -0.87 | 2.4e-03 | Click! |
| chr8_22575948_22576819 | EGR3 | 25568 | 0.121945 | -0.84 | 4.8e-03 | Click! |
| chr8_22571215_22571366 | EGR3 | 20475 | 0.126033 | -0.84 | 4.8e-03 | Click! |
| chr8_22571028_22571179 | EGR3 | 20288 | 0.126164 | -0.83 | 5.8e-03 | Click! |
Activity of the EGR3_EGR2 motif across conditions
Conditions sorted by the z-value of the EGR3_EGR2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
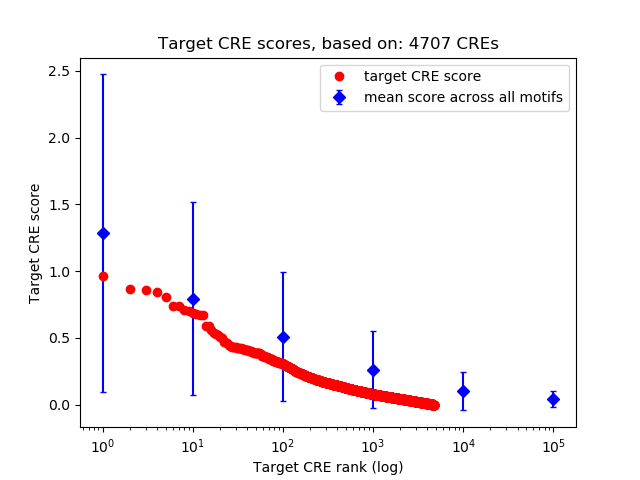
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_113437546_113438608 | 0.97 |
NEUROG2 |
neurogenin 2 |
749 |
0.58 |
| chr13_27540158_27540309 | 0.87 |
USP12-AS1 |
USP12 antisense RNA 1 |
196759 |
0.02 |
| chr4_3709772_3710024 | 0.86 |
ADRA2C |
adrenoceptor alpha 2C |
58177 |
0.14 |
| chr16_88518586_88519370 | 0.84 |
ZFPM1 |
zinc finger protein, FOG family member 1 |
747 |
0.63 |
| chr20_34129990_34130743 | 0.80 |
ERGIC3 |
ERGIC and golgi 3 |
487 |
0.7 |
| chr14_34161217_34161368 | 0.74 |
NPAS3 |
neuronal PAS domain protein 3 |
43155 |
0.21 |
| chr10_127931048_127931232 | 0.74 |
ENSG00000222740 |
. |
96989 |
0.08 |
| chr8_127570759_127570910 | 0.71 |
FAM84B |
family with sequence similarity 84, member B |
196 |
0.83 |
| chr1_220863452_220864325 | 0.70 |
C1orf115 |
chromosome 1 open reading frame 115 |
701 |
0.71 |
| chr4_81228983_81229134 | 0.68 |
C4orf22 |
chromosome 4 open reading frame 22 |
27816 |
0.2 |
| chr11_70963344_70963766 | 0.68 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
68 |
0.98 |
| chr21_47239386_47239537 | 0.67 |
AL592528.1 |
|
16872 |
0.19 |
| chr17_45365957_45366108 | 0.67 |
RP11-290H9.4 |
|
17019 |
0.13 |
| chr14_104182645_104183196 | 0.59 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
775 |
0.34 |
| chr12_26151630_26151781 | 0.59 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
3768 |
0.29 |
| chr18_77265355_77265646 | 0.56 |
AC018445.1 |
Uncharacterized protein |
10557 |
0.27 |
| chr10_128075122_128075273 | 0.54 |
ADAM12 |
ADAM metallopeptidase domain 12 |
1827 |
0.47 |
| chr1_9189681_9190164 | 0.53 |
GPR157 |
G protein-coupled receptor 157 |
693 |
0.66 |
| chr6_131924415_131924566 | 0.52 |
MED23 |
mediator complex subunit 23 |
6845 |
0.18 |
| chr2_239149590_239149855 | 0.51 |
HES6 |
hes family bHLH transcription factor 6 |
419 |
0.75 |
| chr12_132379887_132380588 | 0.50 |
ULK1 |
unc-51 like autophagy activating kinase 1 |
1041 |
0.51 |
| chr18_21762368_21762519 | 0.47 |
OSBPL1A |
oxysterol binding protein-like 1A |
2644 |
0.2 |
| chr5_153990433_153990710 | 0.47 |
ENSG00000264760 |
. |
14939 |
0.21 |
| chr1_161575871_161576361 | 0.46 |
FCGR2C |
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene) |
10773 |
0.12 |
| chr16_88448984_88449499 | 0.45 |
ZNF469 |
zinc finger protein 469 |
44638 |
0.14 |
| chr10_72647360_72647920 | 0.44 |
PCBD1 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
901 |
0.61 |
| chr3_11195342_11196026 | 0.44 |
HRH1 |
histamine receptor H1 |
530 |
0.86 |
| chr20_42143574_42144320 | 0.43 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
781 |
0.59 |
| chr6_149667254_149667417 | 0.43 |
TAB2 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
23709 |
0.17 |
| chr20_17816311_17816543 | 0.43 |
ENSG00000221220 |
. |
6800 |
0.24 |
| chr1_1631163_1631760 | 0.43 |
SLC35E2B |
solute carrier family 35, member E2B |
7294 |
0.11 |
| chr6_170361246_170361397 | 0.42 |
RP11-302L19.1 |
|
116420 |
0.06 |
| chr13_106204547_106204698 | 0.42 |
DAOA-AS1 |
DAOA antisense RNA 1 |
46592 |
0.19 |
| chr11_417004_417487 | 0.42 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
80 |
0.94 |
| chr3_46924910_46925242 | 0.42 |
PTH1R |
parathyroid hormone 1 receptor |
229 |
0.87 |
| chr9_139636321_139636758 | 0.42 |
LCN10 |
lipocalin 10 |
816 |
0.35 |
| chr4_85419435_85420104 | 0.41 |
NKX6-1 |
NK6 homeobox 1 |
166 |
0.97 |
| chr10_105253662_105254118 | 0.41 |
NEURL1 |
neuralized E3 ubiquitin protein ligase 1 |
154 |
0.92 |
| chr5_139283800_139284387 | 0.41 |
NRG2 |
neuregulin 2 |
111 |
0.97 |
| chr17_36031578_36031729 | 0.41 |
DDX52 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 |
28160 |
0.14 |
| chr21_43299631_43300178 | 0.41 |
PRDM15 |
PR domain containing 15 |
313 |
0.82 |
| chr19_17716649_17717262 | 0.40 |
CTD-3149D2.3 |
|
7576 |
0.15 |
| chr12_49739380_49740121 | 0.40 |
DNAJC22 |
DnaJ (Hsp40) homolog, subfamily C, member 22 |
950 |
0.41 |
| chr20_37079281_37079432 | 0.40 |
ENSG00000199266 |
. |
1343 |
0.24 |
| chr11_111217873_111218024 | 0.40 |
ENSG00000264032 |
. |
534 |
0.72 |
| chr19_47524229_47524951 | 0.40 |
NPAS1 |
neuronal PAS domain protein 1 |
447 |
0.8 |
| chr1_38596872_38597023 | 0.39 |
ENSG00000265596 |
. |
42044 |
0.15 |
| chr16_86261645_86261796 | 0.39 |
ENSG00000199949 |
. |
47409 |
0.19 |
| chr9_139462117_139462567 | 0.39 |
RP11-611D20.2 |
|
18760 |
0.09 |
| chr3_183947384_183948074 | 0.39 |
VWA5B2 |
von Willebrand factor A domain containing 5B2 |
488 |
0.65 |
| chr11_64059326_64060001 | 0.39 |
KCNK4 |
potassium channel, subfamily K, member 4 |
181 |
0.83 |
| chr19_18722693_18723176 | 0.39 |
TMEM59L |
transmembrane protein 59-like |
748 |
0.45 |
| chr7_129419053_129419592 | 0.39 |
ENSG00000207691 |
. |
4468 |
0.17 |
| chr11_69209589_69209740 | 0.39 |
MYEOV |
myeloma overexpressed |
148039 |
0.04 |
| chr22_30675082_30675729 | 0.39 |
GATSL3 |
GATS protein-like 3 |
10191 |
0.09 |
| chr3_134034709_134034891 | 0.38 |
AMOTL2 |
angiomotin like 2 |
55954 |
0.13 |
| chr17_79070617_79071314 | 0.38 |
BAIAP2 |
BAI1-associated protein 2 |
414 |
0.74 |
| chr6_17033705_17033856 | 0.37 |
STMND1 |
stathmin domain containing 1 |
68709 |
0.14 |
| chr3_47844781_47845806 | 0.37 |
DHX30 |
DEAH (Asp-Glu-Ala-His) box helicase 30 |
638 |
0.69 |
| chr5_178724831_178724982 | 0.37 |
ADAMTS2 |
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
47525 |
0.17 |
| chr15_80696331_80696627 | 0.36 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
214 |
0.79 |
| chr13_98742061_98742212 | 0.36 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
52680 |
0.14 |
| chr11_68659074_68659228 | 0.36 |
MRPL21 |
mitochondrial ribosomal protein L21 |
12095 |
0.15 |
| chr19_16177822_16178237 | 0.36 |
TPM4 |
tropomyosin 4 |
198 |
0.94 |
| chr3_52039773_52040512 | 0.36 |
RPL29 |
ribosomal protein L29 |
10184 |
0.09 |
| chr14_103673785_103674145 | 0.35 |
ENSG00000239117 |
. |
4442 |
0.24 |
| chr6_147135992_147136143 | 0.35 |
ADGB |
androglobin |
13072 |
0.25 |
| chr22_50422080_50422460 | 0.35 |
IL17REL |
interleukin 17 receptor E-like |
24851 |
0.15 |
| chr19_50355314_50356474 | 0.35 |
PTOV1-AS1 |
PTOV1 antisense RNA 1 |
961 |
0.21 |
| chr8_81477973_81478412 | 0.35 |
ENSG00000223327 |
. |
19816 |
0.2 |
| chr6_5083119_5083270 | 0.35 |
PPP1R3G |
protein phosphatase 1, regulatory subunit 3G |
2526 |
0.31 |
| chr21_46890680_46890997 | 0.34 |
COL18A1 |
collagen, type XVIII, alpha 1 |
15414 |
0.18 |
| chrX_47433764_47434046 | 0.34 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
7807 |
0.13 |
| chr7_128828160_128828607 | 0.34 |
SMO |
smoothened, frizzled family receptor |
330 |
0.84 |
| chr11_63378218_63378369 | 0.34 |
PLA2G16 |
phospholipase A2, group XVI |
2240 |
0.22 |
| chr13_100613171_100613322 | 0.33 |
ZIC5 |
Zic family member 5 |
10917 |
0.19 |
| chr7_73899343_73899494 | 0.33 |
ENSG00000238391 |
. |
2452 |
0.31 |
| chr11_118016566_118017132 | 0.33 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
6686 |
0.16 |
| chr3_25627117_25627268 | 0.33 |
ENSG00000272166 |
. |
30095 |
0.18 |
| chr19_18272446_18272908 | 0.33 |
PIK3R2 |
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
8749 |
0.1 |
| chr11_66103534_66104527 | 0.33 |
RIN1 |
Ras and Rab interactor 1 |
30 |
0.92 |
| chr17_36609068_36609612 | 0.33 |
ENSG00000260833 |
. |
652 |
0.66 |
| chr8_102300306_102300457 | 0.32 |
ENSG00000239115 |
. |
19214 |
0.24 |
| chr16_1203566_1203784 | 0.32 |
CACNA1H |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
63 |
0.95 |
| chr2_106481904_106482055 | 0.32 |
AC009505.2 |
|
8346 |
0.25 |
| chr8_140716343_140716851 | 0.32 |
KCNK9 |
potassium channel, subfamily K, member 9 |
1298 |
0.63 |
| chr17_32907854_32908260 | 0.32 |
TMEM132E |
transmembrane protein 132E |
289 |
0.89 |
| chr1_197924747_197924898 | 0.32 |
LHX9 |
LIM homeobox 9 |
38293 |
0.19 |
| chr4_1003833_1004600 | 0.32 |
FGFRL1 |
fibroblast growth factor receptor-like 1 |
449 |
0.75 |
| chr19_8672060_8672211 | 0.32 |
ADAMTS10 |
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
1048 |
0.45 |
| chrX_3181986_3182137 | 0.31 |
CXorf28 |
chromosome X open reading frame 28 |
7800 |
0.28 |
| chr17_38500621_38501691 | 0.31 |
RARA |
retinoic acid receptor, alpha |
337 |
0.77 |
| chr17_7746157_7747127 | 0.31 |
KDM6B |
lysine (K)-specific demethylase 6B |
1591 |
0.22 |
| chr10_112543415_112543566 | 0.31 |
ENSG00000199364 |
. |
22590 |
0.16 |
| chr12_88375416_88375567 | 0.31 |
C12orf50 |
chromosome 12 open reading frame 50 |
46249 |
0.15 |
| chr8_22026915_22027066 | 0.31 |
BMP1 |
bone morphogenetic protein 1 |
4190 |
0.13 |
| chr1_1014853_1015344 | 0.31 |
RNF223 |
ring finger protein 223 |
5411 |
0.1 |
| chr9_139223916_139224067 | 0.31 |
GPSM1 |
G-protein signaling modulator 1 |
2059 |
0.22 |
| chr6_108105922_108106149 | 0.31 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
12580 |
0.27 |
| chr4_111532636_111532787 | 0.31 |
PITX2 |
paired-like homeodomain 2 |
11496 |
0.28 |
| chr8_143695048_143695766 | 0.30 |
ARC |
activity-regulated cytoskeleton-associated protein |
1426 |
0.34 |
| chr11_12562425_12562576 | 0.30 |
PARVA |
parvin, alpha |
88218 |
0.09 |
| chr14_55574760_55575368 | 0.30 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
15764 |
0.2 |
| chr17_47938739_47938890 | 0.30 |
TAC4 |
tachykinin 4 (hemokinin) |
13435 |
0.16 |
| chr20_62948255_62948712 | 0.30 |
PCMTD2 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
51618 |
0.15 |
| chr16_1202380_1202675 | 0.29 |
CACNA1H |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
714 |
0.52 |
| chr14_69727423_69728222 | 0.29 |
GALNT16 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
814 |
0.7 |
| chr18_46356353_46356504 | 0.29 |
RP11-484L8.1 |
|
4713 |
0.31 |
| chr5_173018079_173018230 | 0.29 |
CTB-33O18.3 |
|
11502 |
0.23 |
| chr2_236516678_236516829 | 0.29 |
ENSG00000221704 |
. |
36178 |
0.19 |
| chr1_42099647_42099798 | 0.29 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
66950 |
0.12 |
| chr12_53297687_53297917 | 0.29 |
KRT8 |
keratin 8 |
370 |
0.82 |
| chr17_45055986_45056489 | 0.29 |
RPRML |
reprimo-like |
377 |
0.82 |
| chr7_16737552_16737703 | 0.29 |
BZW2 |
basic leucine zipper and W2 domains 2 |
9477 |
0.18 |
| chr3_49004665_49004816 | 0.29 |
RP13-131K19.1 |
|
16559 |
0.08 |
| chr21_44144329_44144647 | 0.28 |
AP001627.1 |
|
17380 |
0.2 |
| chr1_203483478_203483842 | 0.28 |
OPTC |
opticin |
18562 |
0.2 |
| chr11_48190887_48191038 | 0.28 |
OR4B1 |
olfactory receptor, family 4, subfamily B, member 1 |
47382 |
0.13 |
| chr13_114516970_114517121 | 0.28 |
GAS6-AS1 |
GAS6 antisense RNA 1 |
1558 |
0.42 |
| chr10_134000631_134000857 | 0.28 |
DPYSL4 |
dihydropyrimidinase-like 4 |
340 |
0.84 |
| chr12_7077943_7078351 | 0.28 |
PHB2 |
prohibitin 2 |
1022 |
0.2 |
| chr17_72754153_72754656 | 0.27 |
SLC9A3R1 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
3738 |
0.12 |
| chr8_42030567_42030894 | 0.27 |
AP3M2 |
adaptor-related protein complex 3, mu 2 subunit |
5923 |
0.17 |
| chr8_93026735_93027023 | 0.27 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
2712 |
0.37 |
| chr14_23357394_23357589 | 0.27 |
REM2 |
RAS (RAD and GEM)-like GTP binding 2 |
5060 |
0.09 |
| chr12_6472663_6473337 | 0.27 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
313 |
0.83 |
| chr3_50232130_50232343 | 0.27 |
GNAT1 |
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
2131 |
0.19 |
| chr13_114898126_114898522 | 0.27 |
RASA3 |
RAS p21 protein activator 3 |
238 |
0.94 |
| chr19_1034551_1034702 | 0.26 |
CNN2 |
calponin 2 |
3490 |
0.09 |
| chr1_15224545_15224696 | 0.26 |
KAZN |
kazrin, periplakin interacting protein |
26006 |
0.26 |
| chr2_38799437_38799781 | 0.26 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
2631 |
0.31 |
| chr21_37572717_37572868 | 0.26 |
ENSG00000265882 |
. |
13344 |
0.14 |
| chr3_156534276_156534726 | 0.26 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
9588 |
0.29 |
| chr19_10531849_10532316 | 0.26 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
751 |
0.43 |
| chr20_62589289_62589587 | 0.26 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
1669 |
0.16 |
| chr1_231410009_231410185 | 0.25 |
ENSG00000222407 |
. |
7173 |
0.17 |
| chr17_17017881_17018380 | 0.25 |
MPRIP |
myosin phosphatase Rho interacting protein |
16850 |
0.15 |
| chr11_12424964_12425115 | 0.25 |
PARVA |
parvin, alpha |
5251 |
0.27 |
| chr13_76223017_76223168 | 0.25 |
LMO7 |
LIM domain 7 |
12633 |
0.18 |
| chr7_100808302_100808702 | 0.25 |
VGF |
VGF nerve growth factor inducible |
107 |
0.93 |
| chr14_105796875_105797044 | 0.25 |
PACS2 |
phosphofurin acidic cluster sorting protein 2 |
5803 |
0.17 |
| chr11_46354061_46354426 | 0.25 |
DGKZ |
diacylglycerol kinase, zeta |
212 |
0.92 |
| chr22_35748735_35749057 | 0.25 |
ENSG00000266320 |
. |
17263 |
0.15 |
| chr1_206654151_206654302 | 0.24 |
IKBKE |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
10388 |
0.14 |
| chr2_219696298_219696449 | 0.24 |
PRKAG3 |
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
141 |
0.94 |
| chr5_55151409_55151585 | 0.24 |
IL31RA |
interleukin 31 receptor A |
2139 |
0.34 |
| chr16_67012908_67013059 | 0.24 |
CES4A |
carboxylesterase 4A |
9509 |
0.11 |
| chr3_71127883_71128284 | 0.24 |
FOXP1 |
forkhead box P1 |
13165 |
0.3 |
| chr8_38261754_38262108 | 0.24 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
0 |
0.96 |
| chr1_180202351_180202502 | 0.24 |
LHX4 |
LIM homeobox 4 |
3005 |
0.29 |
| chr12_133083658_133084311 | 0.24 |
FBRSL1 |
fibrosin-like 1 |
16827 |
0.19 |
| chr2_173139735_173139887 | 0.24 |
ENSG00000238572 |
. |
118963 |
0.05 |
| chr1_156675699_156676565 | 0.24 |
CRABP2 |
cellular retinoic acid binding protein 2 |
524 |
0.6 |
| chr19_45843748_45844099 | 0.24 |
KLC3 |
kinesin light chain 3 |
88 |
0.94 |
| chr7_151574672_151575021 | 0.24 |
PRKAG2-AS1 |
PRKAG2 antisense RNA 1 |
296 |
0.72 |
| chr2_110883713_110883864 | 0.23 |
ENSG00000212091 |
. |
613 |
0.67 |
| chr15_45952586_45952737 | 0.23 |
SQRDL |
sulfide quinone reductase-like (yeast) |
1382 |
0.46 |
| chr6_50818276_50818427 | 0.23 |
TFAP2B |
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
31912 |
0.25 |
| chr1_200004031_200004580 | 0.23 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
7294 |
0.18 |
| chr17_6679303_6679767 | 0.23 |
FBXO39 |
F-box protein 39 |
7 |
0.97 |
| chr6_1620291_1620740 | 0.23 |
FOXC1 |
forkhead box C1 |
9834 |
0.3 |
| chr12_92368232_92368383 | 0.23 |
C12orf79 |
chromosome 12 open reading frame 79 |
162490 |
0.04 |
| chr19_590689_590900 | 0.23 |
HCN2 |
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
901 |
0.37 |
| chr10_13500720_13500871 | 0.23 |
BEND7 |
BEN domain containing 7 |
22278 |
0.16 |
| chr3_120170476_120170627 | 0.23 |
FSTL1 |
follistatin-like 1 |
451 |
0.88 |
| chr9_96720941_96721916 | 0.23 |
BARX1 |
BARX homeobox 1 |
3774 |
0.31 |
| chr14_73392660_73393072 | 0.23 |
DCAF4 |
DDB1 and CUL4 associated factor 4 |
174 |
0.95 |
| chr19_48627309_48627460 | 0.23 |
CTC-453G23.5 |
|
5689 |
0.12 |
| chr9_110925898_110926343 | 0.23 |
ENSG00000222512 |
. |
195089 |
0.03 |
| chr20_31147574_31147725 | 0.23 |
RP11-410N8.3 |
|
1246 |
0.42 |
| chr21_47188043_47188194 | 0.23 |
PRED60 |
|
1887 |
0.41 |
| chr4_176333346_176333497 | 0.23 |
ENSG00000221136 |
. |
60732 |
0.16 |
| chr20_34716861_34717019 | 0.22 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
3574 |
0.2 |
| chr1_181074818_181075147 | 0.22 |
IER5 |
immediate early response 5 |
17344 |
0.22 |
| chr2_20018587_20018738 | 0.22 |
TTC32 |
tetratricopeptide repeat domain 32 |
82740 |
0.1 |
| chr1_206762660_206762811 | 0.22 |
EIF2D |
eukaryotic translation initiation factor 2D |
23068 |
0.13 |
| chr19_4473935_4474715 | 0.22 |
HDGFRP2 |
Hepatoma-derived growth factor-related protein 2 |
564 |
0.54 |
| chr9_21543413_21543564 | 0.22 |
MIR31HG |
MIR31 host gene (non-protein coding) |
16180 |
0.15 |
| chr11_14051655_14051806 | 0.22 |
ENSG00000212365 |
. |
104802 |
0.07 |
| chr1_244080456_244080691 | 0.21 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
66192 |
0.12 |
| chr12_57674022_57674790 | 0.21 |
R3HDM2 |
R3H domain containing 2 |
3371 |
0.16 |
| chr18_14974636_14974787 | 0.21 |
ENSG00000266544 |
. |
115466 |
0.06 |
| chr6_41456635_41457010 | 0.21 |
RP11-328M4.2 |
|
56990 |
0.1 |
| chr1_94080800_94080951 | 0.21 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
1221 |
0.52 |
| chr11_110634298_110634449 | 0.21 |
ARHGAP20 |
Rho GTPase activating protein 20 |
50461 |
0.19 |
| chr2_219748132_219748430 | 0.21 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
1398 |
0.3 |
| chr5_132166329_132166761 | 0.21 |
SHROOM1 |
shroom family member 1 |
45 |
0.96 |
| chr14_103242729_103243285 | 0.21 |
TRAF3 |
TNF receptor-associated factor 3 |
806 |
0.63 |
| chr9_14322498_14322883 | 0.21 |
NFIB |
nuclear factor I/B |
353 |
0.91 |
| chr19_4718186_4718464 | 0.21 |
DPP9 |
dipeptidyl-peptidase 9 |
425 |
0.75 |
| chr17_76919622_76920491 | 0.21 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
1413 |
0.35 |
| chr5_76925321_76925875 | 0.21 |
WDR41 |
WD repeat domain 41 |
9162 |
0.23 |
| chr1_246149947_246150098 | 0.21 |
RP11-83A16.1 |
|
46831 |
0.18 |
| chr1_856648_857113 | 0.21 |
SAMD11 |
sterile alpha motif domain containing 11 |
3380 |
0.14 |
| chr6_146136305_146136901 | 0.21 |
RP11-545I5.3 |
|
169 |
0.88 |
| chr19_40972580_40972926 | 0.21 |
SPTBN4 |
spectrin, beta, non-erythrocytic 4 |
373 |
0.7 |
| chr2_109790847_109791252 | 0.21 |
ENSG00000264934 |
. |
33005 |
0.2 |
| chr5_110584557_110584708 | 0.21 |
AC010468.2 |
|
21155 |
0.19 |
| chr18_55094933_55095260 | 0.21 |
ONECUT2 |
one cut homeobox 2 |
7821 |
0.21 |
| chr12_122158077_122158228 | 0.20 |
TMEM120B |
transmembrane protein 120B |
7451 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.4 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.6 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.1 | 0.2 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.1 | 0.2 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.1 | 0.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.2 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.5 | GO:0010894 | negative regulation of steroid biosynthetic process(GO:0010894) |
| 0.0 | 0.1 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0071883 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0044252 | negative regulation of collagen metabolic process(GO:0010713) negative regulation of multicellular organismal metabolic process(GO:0044252) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0051307 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.2 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.0 | GO:1902339 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0014848 | urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.1 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.0 | 0.1 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.2 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 | 0.1 | GO:1903078 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.0 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:1901374 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.0 | 0.0 | GO:0090189 | regulation of mesonephros development(GO:0061217) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.0 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.0 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0048342 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.1 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0048265 | response to pain(GO:0048265) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.0 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.0 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.4 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.8 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.1 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0080031 | methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.4 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.0 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.0 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.0 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |