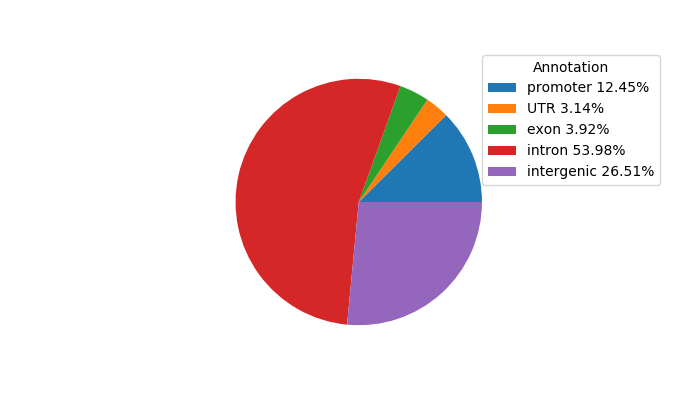
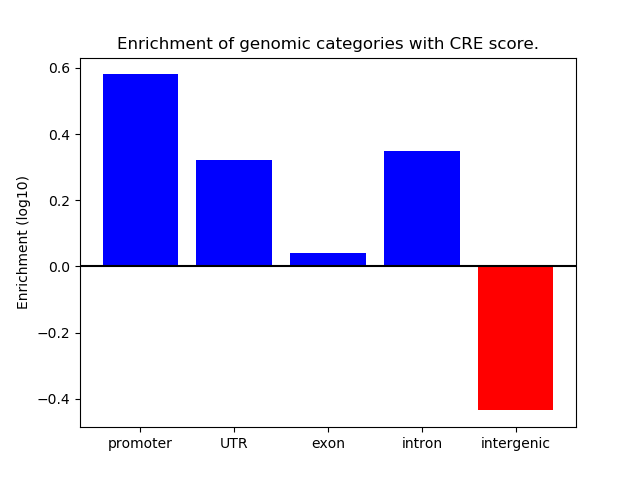
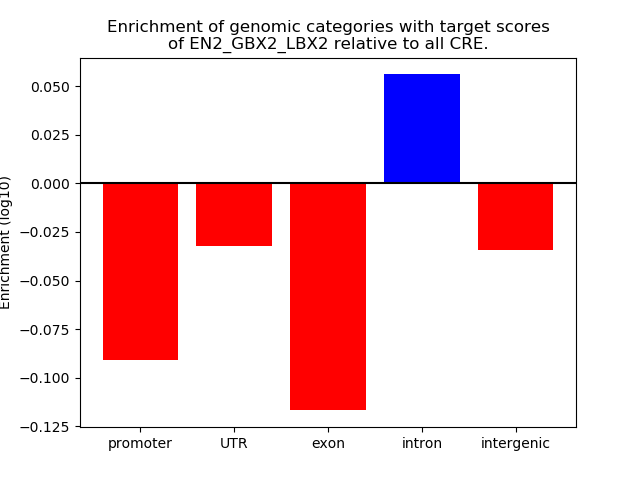
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for EN2_GBX2_LBX2
Z-value: 0.41
Transcription factors associated with EN2_GBX2_LBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EN2
|
ENSG00000164778.4 | engrailed homeobox 2 |
|
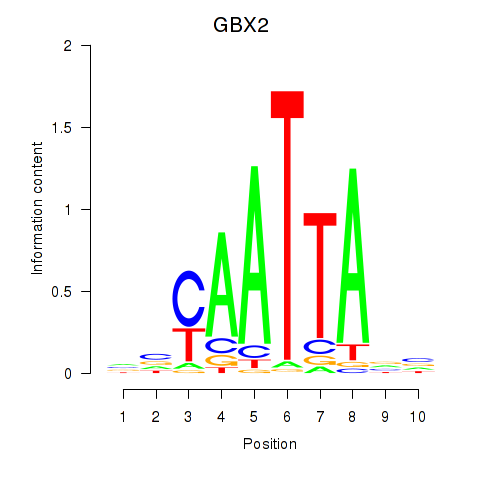
GBX2
|
ENSG00000168505.6 | gastrulation brain homeobox 2 |
|
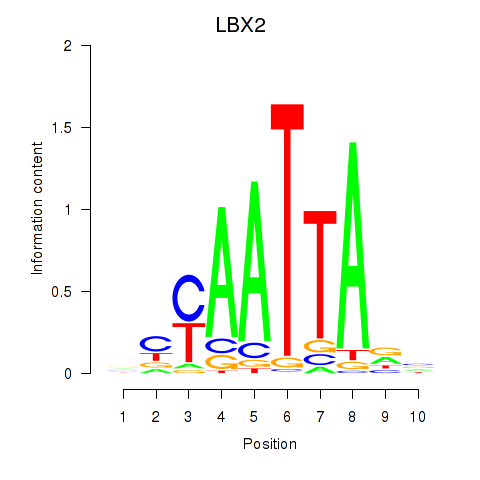
LBX2
|
ENSG00000179528.11 | ladybird homeobox 2 |
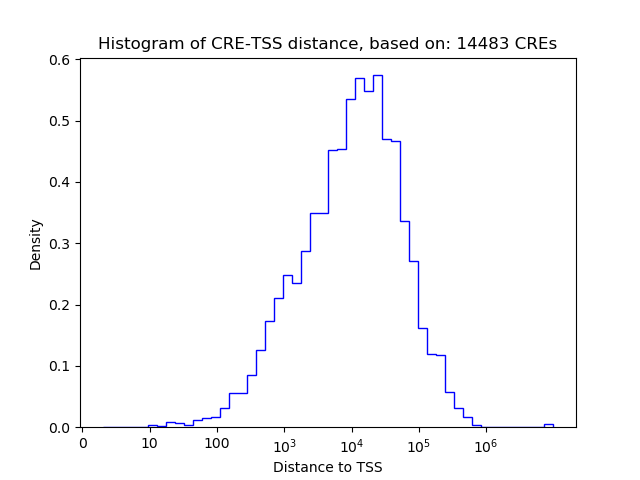
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_155258168_155258329 | EN2 | 7424 | 0.202726 | 0.80 | 9.7e-03 | Click! |
| chr7_155257936_155258087 | EN2 | 7187 | 0.203781 | 0.64 | 6.1e-02 | Click! |
| chr7_155252800_155252951 | EN2 | 2051 | 0.330679 | 0.57 | 1.1e-01 | Click! |
| chr7_155260657_155261013 | EN2 | 10011 | 0.194120 | 0.51 | 1.6e-01 | Click! |
| chr7_155264148_155264299 | EN2 | 13399 | 0.185836 | -0.45 | 2.3e-01 | Click! |
| chr2_237088836_237088987 | GBX2 | 11899 | 0.144788 | -0.60 | 8.7e-02 | Click! |
| chr2_237088666_237088817 | GBX2 | 11729 | 0.145069 | -0.60 | 9.0e-02 | Click! |
| chr2_237081005_237081156 | GBX2 | 4068 | 0.175730 | 0.45 | 2.2e-01 | Click! |
| chr2_237077629_237077780 | GBX2 | 692 | 0.570341 | -0.38 | 3.2e-01 | Click! |
| chr2_237087329_237087480 | GBX2 | 10392 | 0.147383 | 0.33 | 3.8e-01 | Click! |
| chr2_74726899_74727050 | LBX2 | 131 | 0.757860 | 0.58 | 1.0e-01 | Click! |
| chr2_74731286_74731437 | LBX2 | 918 | 0.240961 | -0.51 | 1.7e-01 | Click! |
| chr2_74727069_74727321 | LBX2 | 352 | 0.531282 | -0.44 | 2.4e-01 | Click! |
| chr2_74732516_74733402 | LBX2 | 2516 | 0.086291 | -0.40 | 2.8e-01 | Click! |
| chr2_74732343_74732494 | LBX2 | 1975 | 0.105454 | -0.40 | 2.9e-01 | Click! |
Activity of the EN2_GBX2_LBX2 motif across conditions
Conditions sorted by the z-value of the EN2_GBX2_LBX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
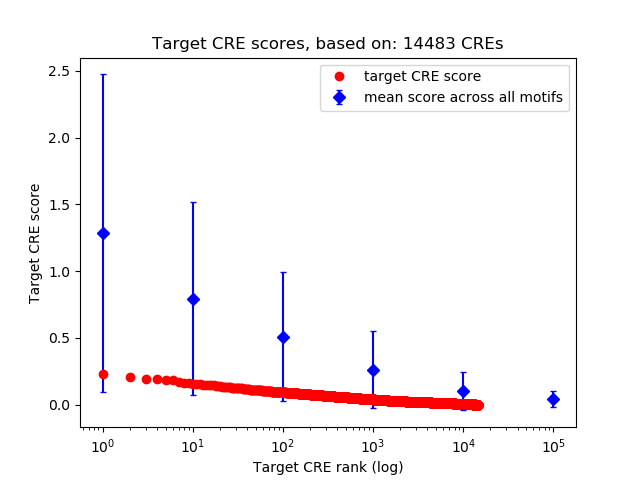
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_101394347_101394839 | 0.23 |
SLC30A7 |
solute carrier family 30 (zinc transporter), member 7 |
32783 |
0.12 |
| chr21_26937814_26938070 | 0.21 |
ENSG00000234883 |
. |
8350 |
0.19 |
| chr16_73092870_73093114 | 0.20 |
ZFHX3 |
zinc finger homeobox 3 |
605 |
0.79 |
| chr8_49862673_49862887 | 0.20 |
SNAI2 |
snail family zinc finger 2 |
28481 |
0.25 |
| chr9_117142014_117142165 | 0.19 |
AKNA |
AT-hook transcription factor |
2174 |
0.33 |
| chr13_46893880_46894070 | 0.18 |
ENSG00000223336 |
. |
54750 |
0.1 |
| chr1_169672251_169672700 | 0.17 |
SELL |
selectin L |
8364 |
0.2 |
| chr3_186237166_186237573 | 0.16 |
CRYGS |
crystallin, gamma S |
24871 |
0.16 |
| chr6_74169990_74170157 | 0.16 |
MTO1 |
mitochondrial tRNA translation optimization 1 |
1228 |
0.27 |
| chr5_100190438_100190674 | 0.16 |
ENSG00000221263 |
. |
38287 |
0.19 |
| chr1_172666866_172667065 | 0.15 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
38807 |
0.19 |
| chr21_36305039_36305387 | 0.15 |
RUNX1 |
runt-related transcription factor 1 |
43126 |
0.21 |
| chr15_39915311_39915462 | 0.15 |
FSIP1 |
fibrous sheath interacting protein 1 |
22259 |
0.18 |
| chr2_30557961_30558163 | 0.15 |
ENSG00000221377 |
. |
97252 |
0.07 |
| chr2_198100640_198101238 | 0.15 |
ANKRD44 |
ankyrin repeat domain 44 |
38177 |
0.14 |
| chr12_68382310_68382485 | 0.15 |
IFNG-AS1 |
IFNG antisense RNA 1 |
828 |
0.76 |
| chr18_77128932_77129101 | 0.15 |
RP11-800A18.4 |
|
25044 |
0.17 |
| chr19_47948776_47948927 | 0.14 |
MEIS3 |
Meis homeobox 3 |
26071 |
0.11 |
| chr22_39714814_39715056 | 0.14 |
RPL3 |
ribosomal protein L3 |
181 |
0.48 |
| chr10_11201909_11202161 | 0.14 |
CELF2 |
CUGBP, Elav-like family member 2 |
4958 |
0.24 |
| chr10_11248462_11248734 | 0.14 |
RP3-323N1.2 |
|
35259 |
0.18 |
| chr16_74620197_74620348 | 0.13 |
GLG1 |
golgi glycoprotein 1 |
20720 |
0.2 |
| chr5_76939960_76940111 | 0.13 |
OTP |
orthopedia homeobox |
4522 |
0.27 |
| chr3_16883717_16883868 | 0.13 |
PLCL2 |
phospholipase C-like 2 |
42660 |
0.19 |
| chr5_133425812_133426164 | 0.13 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
24414 |
0.2 |
| chr4_109037661_109037882 | 0.13 |
LEF1 |
lymphoid enhancer-binding factor 1 |
49686 |
0.14 |
| chr1_3806778_3806979 | 0.13 |
ENSG00000264428 |
. |
6250 |
0.16 |
| chr7_142024322_142024543 | 0.12 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
34823 |
0.18 |
| chr2_204726088_204726239 | 0.12 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
6346 |
0.27 |
| chr17_38376031_38376425 | 0.12 |
WIPF2 |
WAS/WASL interacting protein family, member 2 |
124 |
0.95 |
| chr11_6425333_6425495 | 0.12 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
480 |
0.74 |
| chr2_197039401_197039716 | 0.12 |
STK17B |
serine/threonine kinase 17b |
1493 |
0.42 |
| chr3_71516692_71516843 | 0.12 |
ENSG00000221264 |
. |
74473 |
0.1 |
| chr3_16971345_16971496 | 0.12 |
PLCL2 |
phospholipase C-like 2 |
3162 |
0.28 |
| chr3_32376993_32377254 | 0.12 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
56040 |
0.12 |
| chrX_118821503_118821974 | 0.12 |
SEPT6 |
septin 6 |
5054 |
0.2 |
| chr9_95786987_95787346 | 0.12 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
9824 |
0.19 |
| chr17_71199413_71199564 | 0.12 |
COG1 |
component of oligomeric golgi complex 1 |
2266 |
0.24 |
| chrX_117644955_117645193 | 0.12 |
DOCK11 |
dedicator of cytokinesis 11 |
15202 |
0.24 |
| chr7_100140293_100140446 | 0.12 |
AGFG2 |
ArfGAP with FG repeats 2 |
3487 |
0.11 |
| chr15_70554588_70554739 | 0.11 |
ENSG00000200216 |
. |
69088 |
0.13 |
| chr3_43064995_43065183 | 0.11 |
FAM198A |
family with sequence similarity 198, member A |
44049 |
0.13 |
| chr13_108979091_108979372 | 0.11 |
ENSG00000223177 |
. |
25552 |
0.22 |
| chr20_47334105_47334366 | 0.11 |
ENSG00000251876 |
. |
21750 |
0.26 |
| chr16_16193929_16194080 | 0.11 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
90291 |
0.07 |
| chr2_18003162_18003313 | 0.11 |
MSGN1 |
mesogenin 1 |
5474 |
0.22 |
| chr5_109032486_109032637 | 0.11 |
ENSG00000202512 |
. |
2974 |
0.27 |
| chr12_68530648_68530810 | 0.11 |
IFNG |
interferon, gamma |
22798 |
0.24 |
| chr6_139616194_139616345 | 0.11 |
TXLNB |
taxilin beta |
2993 |
0.28 |
| chr13_20985266_20985502 | 0.11 |
ENSG00000263978 |
. |
22601 |
0.23 |
| chr11_128589269_128589420 | 0.11 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
23426 |
0.17 |
| chr1_41810525_41810676 | 0.11 |
FOXO6 |
forkhead box O6 |
16994 |
0.21 |
| chr17_17084049_17084292 | 0.11 |
MPRIP |
myosin phosphatase Rho interacting protein |
1298 |
0.29 |
| chr6_15078179_15078392 | 0.11 |
ENSG00000242989 |
. |
34914 |
0.21 |
| chr10_111845210_111845446 | 0.11 |
ADD3 |
adducin 3 (gamma) |
77606 |
0.09 |
| chr18_5158274_5158519 | 0.11 |
C18orf42 |
chromosome 18 open reading frame 42 |
38828 |
0.21 |
| chr1_200987046_200987749 | 0.11 |
KIF21B |
kinesin family member 21B |
5139 |
0.21 |
| chr11_64541210_64541478 | 0.11 |
SF1 |
splicing factor 1 |
3888 |
0.14 |
| chr9_36218264_36218537 | 0.11 |
CLTA |
clathrin, light chain A |
27440 |
0.14 |
| chr5_133881433_133881702 | 0.11 |
JADE2 |
jade family PHD finger 2 |
19240 |
0.15 |
| chr8_52913099_52913333 | 0.11 |
RP11-110G21.1 |
|
100922 |
0.07 |
| chr20_31116051_31116202 | 0.11 |
C20orf112 |
chromosome 20 open reading frame 112 |
7469 |
0.18 |
| chr2_8615200_8615353 | 0.11 |
AC011747.7 |
|
200620 |
0.03 |
| chr13_51487581_51487732 | 0.10 |
RNASEH2B-AS1 |
RNASEH2B antisense RNA 1 |
2808 |
0.26 |
| chr13_32998493_32998691 | 0.10 |
N4BP2L1 |
NEDD4 binding protein 2-like 1 |
3559 |
0.24 |
| chr3_81913343_81913495 | 0.10 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
102107 |
0.09 |
| chr2_111506043_111506194 | 0.10 |
ACOXL |
acyl-CoA oxidase-like |
15686 |
0.25 |
| chr17_47872989_47873209 | 0.10 |
KAT7 |
K(lysine) acetyltransferase 7 |
6084 |
0.16 |
| chr14_93315070_93315221 | 0.10 |
GOLGA5 |
golgin A5 |
15674 |
0.24 |
| chr1_60036490_60036813 | 0.10 |
FGGY |
FGGY carbohydrate kinase domain containing |
17061 |
0.29 |
| chr12_93827062_93827301 | 0.10 |
UBE2N |
ubiquitin-conjugating enzyme E2N |
7851 |
0.16 |
| chr2_182171124_182171288 | 0.10 |
ENSG00000266705 |
. |
827 |
0.77 |
| chr17_42579880_42580174 | 0.10 |
GPATCH8 |
G patch domain containing 8 |
732 |
0.66 |
| chr1_235439552_235439703 | 0.10 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
51038 |
0.11 |
| chr12_2472562_2472713 | 0.10 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
93695 |
0.08 |
| chr10_61631584_61631735 | 0.10 |
CCDC6 |
coiled-coil domain containing 6 |
34755 |
0.21 |
| chr4_140971490_140971641 | 0.10 |
RP11-392B6.1 |
|
77604 |
0.1 |
| chr14_66460757_66460908 | 0.10 |
CTD-2014B16.3 |
Uncharacterized protein |
10409 |
0.25 |
| chr3_135908620_135908771 | 0.10 |
MSL2 |
male-specific lethal 2 homolog (Drosophila) |
4701 |
0.3 |
| chr15_52338744_52338984 | 0.10 |
RP11-430B1.1 |
|
12960 |
0.15 |
| chr13_113840073_113840224 | 0.10 |
RP11-98F14.11 |
|
20651 |
0.11 |
| chr12_89632258_89632472 | 0.10 |
ENSG00000238302 |
. |
43697 |
0.19 |
| chr8_66873049_66873243 | 0.10 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
60649 |
0.14 |
| chr18_11852927_11853152 | 0.10 |
RP11-78A19.3 |
|
289 |
0.82 |
| chr2_144014874_144015136 | 0.10 |
RP11-190J23.1 |
|
85264 |
0.1 |
| chr1_211913474_211913689 | 0.10 |
ENSG00000241395 |
. |
52167 |
0.1 |
| chr3_128836654_128836805 | 0.10 |
RAB43 |
RAB43, member RAS oncogene family |
3895 |
0.15 |
| chr17_40574094_40574415 | 0.10 |
PTRF |
polymerase I and transcript release factor |
1281 |
0.3 |
| chr14_66304578_66304729 | 0.10 |
CTD-2014B16.3 |
Uncharacterized protein |
166588 |
0.04 |
| chr2_26018392_26018619 | 0.10 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
15327 |
0.23 |
| chr20_44634089_44634268 | 0.10 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
3369 |
0.16 |
| chr6_11276274_11276599 | 0.10 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
43521 |
0.17 |
| chr1_100905257_100905795 | 0.10 |
RP5-837M10.4 |
|
46027 |
0.13 |
| chr12_27107136_27107312 | 0.09 |
FGFR1OP2 |
FGFR1 oncogene partner 2 |
15791 |
0.13 |
| chr1_235438349_235438790 | 0.09 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
52096 |
0.11 |
| chr19_8639084_8639286 | 0.09 |
MYO1F |
myosin IF |
3137 |
0.16 |
| chr1_166133959_166134203 | 0.09 |
RP11-9L18.3 |
|
251 |
0.9 |
| chr3_151932265_151932416 | 0.09 |
MBNL1 |
muscleblind-like splicing regulator 1 |
53489 |
0.14 |
| chr16_79652126_79652340 | 0.09 |
MAF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
17622 |
0.26 |
| chr14_50530285_50530492 | 0.09 |
ENSG00000201358 |
. |
4898 |
0.18 |
| chr6_167550535_167550692 | 0.09 |
TCP10L2 |
t-complex 10-like 2 |
12355 |
0.17 |
| chrX_70325744_70326037 | 0.09 |
CXorf65 |
chromosome X open reading frame 65 |
565 |
0.6 |
| chr9_70851374_70851561 | 0.09 |
CBWD3 |
COBW domain containing 3 |
4930 |
0.19 |
| chr11_118097693_118098499 | 0.09 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
2287 |
0.23 |
| chr5_169708109_169708260 | 0.09 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
13853 |
0.22 |
| chr10_121543359_121543582 | 0.09 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
2331 |
0.32 |
| chr9_92078215_92078535 | 0.09 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
7507 |
0.25 |
| chr9_71633765_71634208 | 0.09 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
4947 |
0.24 |
| chr6_42740405_42740590 | 0.09 |
GLTSCR1L |
GLTSCR1-like |
9268 |
0.17 |
| chr1_193428730_193429035 | 0.09 |
ENSG00000252241 |
. |
272192 |
0.01 |
| chr5_61873952_61874158 | 0.09 |
LRRC70 |
leucine rich repeat containing 70 |
507 |
0.52 |
| chr8_29945094_29945295 | 0.09 |
TMEM66 |
transmembrane protein 66 |
4471 |
0.16 |
| chr1_173487659_173487906 | 0.09 |
RP3-436N22.3 |
|
36608 |
0.14 |
| chr7_21469388_21469667 | 0.09 |
SP4 |
Sp4 transcription factor |
1866 |
0.34 |
| chr22_21922789_21923120 | 0.09 |
UBE2L3 |
ubiquitin-conjugating enzyme E2L 3 |
928 |
0.38 |
| chr12_111613238_111613505 | 0.09 |
CUX2 |
cut-like homeobox 2 |
76121 |
0.1 |
| chr20_57723034_57723340 | 0.09 |
ZNF831 |
zinc finger protein 831 |
42888 |
0.16 |
| chr1_42123736_42123959 | 0.09 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
42825 |
0.18 |
| chr2_198072900_198073187 | 0.09 |
ANKRD44 |
ankyrin repeat domain 44 |
10281 |
0.21 |
| chr16_3058535_3058724 | 0.09 |
LA16c-380H5.2 |
|
3527 |
0.07 |
| chr1_42273293_42273444 | 0.09 |
ENSG00000264896 |
. |
48556 |
0.18 |
| chr6_42740244_42740405 | 0.09 |
GLTSCR1L |
GLTSCR1-like |
9441 |
0.17 |
| chr3_98273092_98273322 | 0.09 |
GPR15 |
G protein-coupled receptor 15 |
22464 |
0.13 |
| chr6_42409736_42409944 | 0.09 |
TRERF1 |
transcriptional regulating factor 1 |
9159 |
0.21 |
| chr1_235587141_235587347 | 0.09 |
TBCE-AS1 |
TBCE antisense RNA 1 |
3054 |
0.29 |
| chr16_3125440_3125843 | 0.09 |
ENSG00000252561 |
. |
6117 |
0.06 |
| chr1_151869347_151869629 | 0.09 |
THEM4 |
thioesterase superfamily member 4 |
12625 |
0.12 |
| chr2_198057877_198058189 | 0.09 |
AC013264.2 |
|
4682 |
0.23 |
| chrX_149646217_149646416 | 0.09 |
MAMLD1 |
mastermind-like domain containing 1 |
7036 |
0.24 |
| chr1_158788831_158789034 | 0.09 |
MNDA |
myeloid cell nuclear differentiation antigen |
12175 |
0.17 |
| chr8_103117650_103117985 | 0.09 |
NCALD |
neurocalcin delta |
18361 |
0.2 |
| chr9_135382524_135382903 | 0.09 |
BARHL1 |
BarH-like homeobox 1 |
74859 |
0.09 |
| chr19_47438297_47438633 | 0.09 |
ENSG00000252071 |
. |
13081 |
0.16 |
| chr22_36550343_36550563 | 0.09 |
APOL3 |
apolipoprotein L, 3 |
6378 |
0.24 |
| chr11_20179253_20179404 | 0.09 |
DBX1 |
developing brain homeobox 1 |
2542 |
0.34 |
| chr7_25560467_25560618 | 0.09 |
ENSG00000222101 |
. |
49746 |
0.19 |
| chr5_75600048_75600202 | 0.09 |
RP11-466P24.6 |
|
7162 |
0.3 |
| chr2_114189862_114190032 | 0.09 |
CBWD2 |
COBW domain containing 2 |
5321 |
0.16 |
| chr1_110333910_110334183 | 0.09 |
EPS8L3 |
EPS8-like 3 |
27397 |
0.13 |
| chr7_143007723_143007933 | 0.09 |
CLCN1 |
chloride channel, voltage-sensitive 1 |
5391 |
0.1 |
| chr5_67552449_67552681 | 0.09 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
16839 |
0.26 |
| chr18_56438172_56438323 | 0.08 |
RP11-126O1.4 |
|
18285 |
0.15 |
| chr19_5906272_5906423 | 0.08 |
VMAC |
vimentin-type intermediate filament associated coiled-coil protein |
1478 |
0.18 |
| chr5_179433301_179433452 | 0.08 |
ENSG00000198995 |
. |
9021 |
0.19 |
| chr1_42267028_42267179 | 0.08 |
ENSG00000264896 |
. |
42291 |
0.19 |
| chr4_140724600_140724880 | 0.08 |
ENSG00000252233 |
. |
9153 |
0.26 |
| chr19_19185104_19185255 | 0.08 |
SLC25A42 |
solute carrier family 25, member 42 |
10371 |
0.13 |
| chr15_40342145_40342411 | 0.08 |
SRP14 |
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
10889 |
0.15 |
| chr21_36367069_36367374 | 0.08 |
RUNX1 |
runt-related transcription factor 1 |
54241 |
0.18 |
| chr20_50077608_50077759 | 0.08 |
ENSG00000266761 |
. |
8169 |
0.27 |
| chr19_41811849_41812000 | 0.08 |
CCDC97 |
coiled-coil domain containing 97 |
4170 |
0.13 |
| chr18_43266469_43266620 | 0.08 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
20437 |
0.17 |
| chrX_42259008_42259366 | 0.08 |
ENSG00000206896 |
. |
225549 |
0.02 |
| chr22_26988789_26989025 | 0.08 |
TPST2 |
tyrosylprotein sulfotransferase 2 |
2780 |
0.23 |
| chr8_56756381_56756653 | 0.08 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
35855 |
0.12 |
| chr12_2349139_2349290 | 0.08 |
CACNA1C-AS4 |
CACNA1C antisense RNA 4 |
16569 |
0.22 |
| chr12_104865091_104865422 | 0.08 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
14477 |
0.26 |
| chr14_60761646_60761856 | 0.08 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
38271 |
0.17 |
| chr1_117058467_117058618 | 0.08 |
CD58 |
CD58 molecule |
28670 |
0.15 |
| chr10_49719976_49720127 | 0.08 |
ARHGAP22-IT1 |
ARHGAP22 intronic transcript 1 (non-protein coding) |
419 |
0.87 |
| chr3_107820810_107821187 | 0.08 |
CD47 |
CD47 molecule |
11126 |
0.3 |
| chr5_118710365_118710516 | 0.08 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
18710 |
0.2 |
| chr2_37192955_37193106 | 0.08 |
STRN |
striatin, calmodulin binding protein |
576 |
0.74 |
| chr7_135623902_135624121 | 0.08 |
LUZP6 |
leucine zipper protein 6 |
11813 |
0.14 |
| chr4_89526439_89526704 | 0.08 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
374 |
0.86 |
| chr1_26441433_26441584 | 0.08 |
PDIK1L |
PDLIM1 interacting kinase 1 like |
3168 |
0.15 |
| chr11_72866229_72866512 | 0.08 |
FCHSD2 |
FCH and double SH3 domains 2 |
13064 |
0.2 |
| chr7_21004042_21004193 | 0.08 |
SP8 |
Sp8 transcription factor |
177612 |
0.03 |
| chr5_96345721_96346249 | 0.08 |
ENSG00000200884 |
. |
47524 |
0.12 |
| chr10_90144841_90145001 | 0.08 |
RNLS |
renalase, FAD-dependent amine oxidase |
198026 |
0.02 |
| chr7_102072492_102072712 | 0.08 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
951 |
0.34 |
| chr10_6642170_6642420 | 0.08 |
PRKCQ |
protein kinase C, theta |
20032 |
0.28 |
| chr6_108139065_108139216 | 0.08 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
6376 |
0.29 |
| chr1_28526158_28526348 | 0.08 |
AL353354.2 |
|
817 |
0.34 |
| chr6_13686993_13687323 | 0.08 |
RANBP9 |
RAN binding protein 9 |
22 |
0.98 |
| chr6_122733810_122733961 | 0.08 |
HSF2 |
heat shock transcription factor 2 |
9416 |
0.25 |
| chr4_42634615_42634846 | 0.08 |
ATP8A1 |
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
24380 |
0.21 |
| chr9_92022718_92022899 | 0.08 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
1967 |
0.41 |
| chr2_143885609_143886070 | 0.08 |
ARHGAP15 |
Rho GTPase activating protein 15 |
1044 |
0.64 |
| chr2_143914477_143914920 | 0.08 |
RP11-190J23.1 |
|
15043 |
0.24 |
| chr10_11155205_11156000 | 0.08 |
CELF2-AS2 |
CELF2 antisense RNA 2 |
15166 |
0.21 |
| chr9_70495481_70495632 | 0.08 |
CBWD5 |
COBW domain containing 5 |
5310 |
0.29 |
| chr1_108624991_108625142 | 0.08 |
ENSG00000264753 |
. |
63673 |
0.12 |
| chr8_19552777_19552928 | 0.08 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
12580 |
0.3 |
| chr11_128348480_128348737 | 0.08 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
26681 |
0.22 |
| chr22_40322800_40322970 | 0.08 |
GRAP2 |
GRB2-related adaptor protein 2 |
244 |
0.92 |
| chr12_55372614_55372982 | 0.08 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
2824 |
0.31 |
| chr16_50802191_50802342 | 0.08 |
CYLD |
cylindromatosis (turban tumor syndrome) |
18806 |
0.11 |
| chr12_47600641_47601211 | 0.08 |
PCED1B |
PC-esterase domain containing 1B |
9126 |
0.22 |
| chr11_13944070_13944221 | 0.08 |
ENSG00000201856 |
. |
15008 |
0.29 |
| chr8_59478164_59478344 | 0.08 |
SDCBP |
syndecan binding protein (syntenin) |
679 |
0.77 |
| chrX_102787503_102787654 | 0.08 |
RAB40A |
RAB40A, member RAS oncogene family |
13161 |
0.15 |
| chr16_85971060_85971235 | 0.08 |
IRF8 |
interferon regulatory factor 8 |
23228 |
0.22 |
| chr1_89992034_89992185 | 0.08 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
1647 |
0.46 |
| chr12_21766880_21767202 | 0.08 |
GYS2 |
glycogen synthase 2 (liver) |
9260 |
0.2 |
| chr3_33102387_33102538 | 0.08 |
GLB1 |
galactosidase, beta 1 |
35822 |
0.13 |
| chrX_19766831_19767108 | 0.08 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
1148 |
0.65 |
| chr2_182100905_182101073 | 0.08 |
ENSG00000266705 |
. |
69390 |
0.13 |
| chr11_64537353_64537738 | 0.08 |
SF1 |
splicing factor 1 |
1002 |
0.4 |
| chr10_6096471_6096850 | 0.08 |
IL2RA |
interleukin 2 receptor, alpha |
7593 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0097300 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.0 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:1901985 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.0 | GO:0051532 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0002666 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.0 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.0 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.0 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |