Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
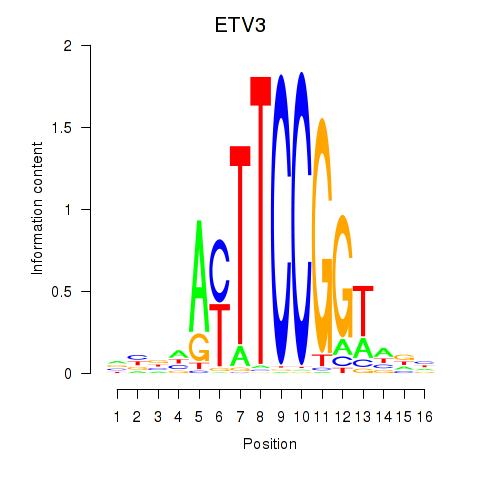
Results for ETV3
Z-value: 0.56
Transcription factors associated with ETV3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV3
|
ENSG00000117036.7 | ETS variant transcription factor 3 |
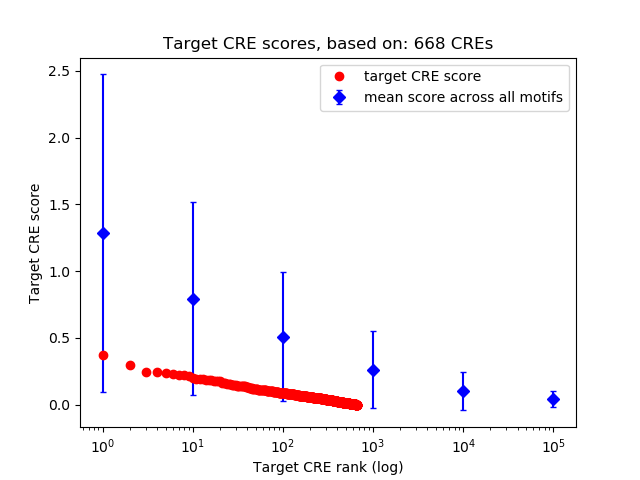
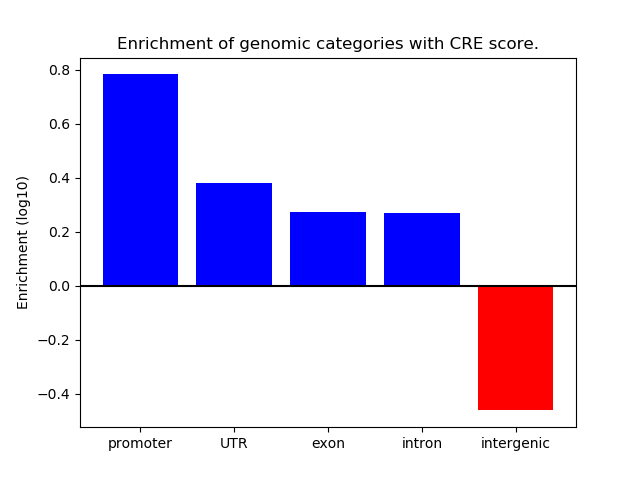
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_157103928_157104177 | ETV3 | 4107 | 0.257990 | -0.90 | 8.7e-04 | Click! |
| chr1_157104246_157104397 | ETV3 | 3838 | 0.264218 | -0.89 | 1.4e-03 | Click! |
| chr1_157102658_157102809 | ETV3 | 5426 | 0.238179 | -0.88 | 1.7e-03 | Click! |
| chr1_157104640_157104791 | ETV3 | 3444 | 0.275656 | -0.86 | 3.3e-03 | Click! |
| chr1_157106662_157106860 | ETV3 | 1398 | 0.478275 | -0.83 | 6.0e-03 | Click! |
Activity of the ETV3 motif across conditions
Conditions sorted by the z-value of the ETV3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
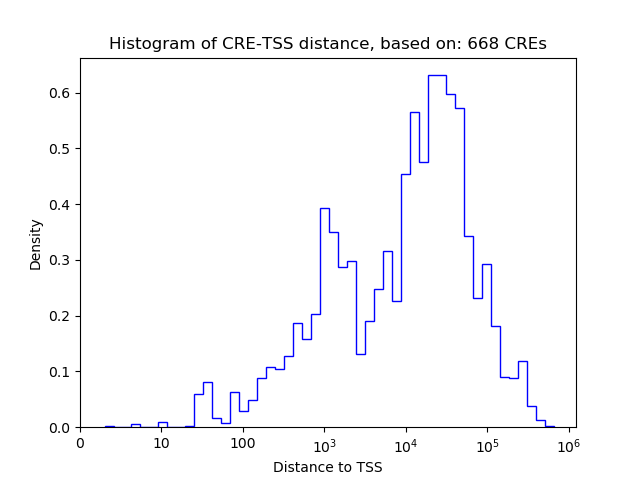
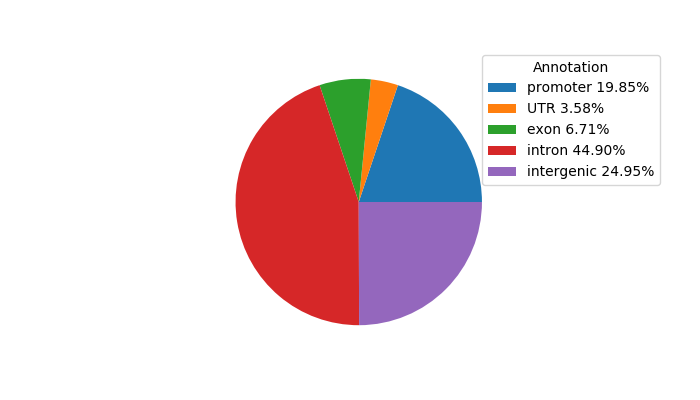
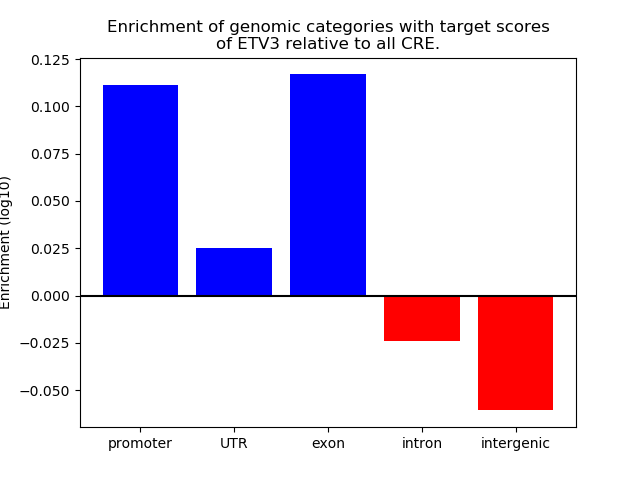
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_33522924_33523075 | 0.37 |
NRP1 |
neuropilin 1 |
20510 |
0.2 |
| chr2_102080787_102081074 | 0.29 |
RFX8 |
RFX family member 8, lacking RFX DNA binding domain |
9807 |
0.26 |
| chr22_30604075_30604921 | 0.25 |
RP3-438O4.4 |
|
1400 |
0.33 |
| chr8_124693439_124693590 | 0.24 |
KLHL38 |
kelch-like family member 38 |
28324 |
0.16 |
| chr4_185292361_185292546 | 0.24 |
IRF2 |
interferon regulatory factor 2 |
47291 |
0.14 |
| chr4_189329906_189330057 | 0.23 |
RP11-366H4.3 |
|
265448 |
0.02 |
| chr8_81938860_81939124 | 0.22 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
85311 |
0.1 |
| chr5_168549003_168549154 | 0.22 |
CTB-174D11.1 |
|
91635 |
0.09 |
| chr7_101361818_101362221 | 0.21 |
MYL10 |
myosin, light chain 10, regulatory |
89443 |
0.08 |
| chr1_20062070_20062221 | 0.20 |
TMCO4 |
transmembrane and coiled-coil domains 4 |
63692 |
0.08 |
| chr1_78468694_78468845 | 0.19 |
RP11-386I14.4 |
|
1469 |
0.3 |
| chr18_12306533_12306684 | 0.19 |
TUBB6 |
tubulin, beta 6 class V |
1060 |
0.47 |
| chr15_59587948_59588157 | 0.19 |
RP11-429D19.1 |
|
24691 |
0.13 |
| chr7_97958337_97958488 | 0.19 |
RP11-307C18.1 |
|
6247 |
0.2 |
| chr5_71579248_71579540 | 0.19 |
ENSG00000244748 |
. |
31121 |
0.16 |
| chr1_17914051_17914202 | 0.18 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
785 |
0.75 |
| chr9_38437379_38437604 | 0.18 |
IGFBPL1 |
insulin-like growth factor binding protein-like 1 |
13047 |
0.24 |
| chr13_111856507_111856685 | 0.18 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
1040 |
0.61 |
| chr14_53250618_53250769 | 0.18 |
ENSG00000264700 |
. |
1602 |
0.32 |
| chr2_165245042_165245193 | 0.18 |
ENSG00000264074 |
. |
34918 |
0.24 |
| chr12_15784371_15784522 | 0.16 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
30762 |
0.18 |
| chr5_167115956_167116107 | 0.16 |
CTB-78F1.1 |
|
28545 |
0.19 |
| chr3_156526171_156526322 | 0.16 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
17843 |
0.26 |
| chr16_24991295_24991446 | 0.16 |
ARHGAP17 |
Rho GTPase activating protein 17 |
35282 |
0.2 |
| chr11_130431461_130431612 | 0.16 |
C11orf44 |
chromosome 11 open reading frame 44 |
111315 |
0.06 |
| chr17_72968449_72968753 | 0.15 |
HID1 |
HID1 domain containing |
210 |
0.85 |
| chr4_6273239_6273390 | 0.15 |
WFS1 |
Wolfram syndrome 1 (wolframin) |
1737 |
0.47 |
| chr7_100777693_100777872 | 0.15 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
7403 |
0.1 |
| chr21_39228429_39228580 | 0.15 |
KCNJ6 |
potassium inwardly-rectifying channel, subfamily J, member 6 |
57053 |
0.14 |
| chr14_68467342_68467493 | 0.15 |
CTD-2566J3.1 |
|
129496 |
0.05 |
| chr10_5643353_5643504 | 0.14 |
ENSG00000240577 |
. |
29998 |
0.14 |
| chr8_1941875_1942026 | 0.14 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
19906 |
0.23 |
| chr2_121947507_121947892 | 0.14 |
TFCP2L1 |
transcription factor CP2-like 1 |
95084 |
0.08 |
| chr19_1253556_1254162 | 0.14 |
MIDN |
midnolin |
2238 |
0.14 |
| chrX_119683710_119683861 | 0.14 |
ENSG00000272179 |
. |
5615 |
0.2 |
| chr20_52517473_52517681 | 0.14 |
AC005220.3 |
|
39122 |
0.2 |
| chr5_152825461_152825612 | 0.14 |
GRIA1 |
glutamate receptor, ionotropic, AMPA 1 |
44570 |
0.21 |
| chr16_87849935_87850253 | 0.14 |
RP4-536B24.2 |
|
20044 |
0.16 |
| chr11_100680065_100680216 | 0.13 |
ENSG00000265208 |
. |
14968 |
0.22 |
| chr22_43411349_43411733 | 0.13 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
390 |
0.86 |
| chr2_217400437_217400588 | 0.13 |
RPL37A |
ribosomal protein L37a |
36649 |
0.12 |
| chr16_27378848_27379069 | 0.13 |
IL4R |
interleukin 4 receptor |
14962 |
0.19 |
| chr3_188680922_188681073 | 0.12 |
TPRG1 |
tumor protein p63 regulated 1 |
15994 |
0.3 |
| chr7_151006676_151006827 | 0.12 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
31769 |
0.1 |
| chr19_19510818_19511425 | 0.12 |
CTB-184G21.3 |
|
2291 |
0.22 |
| chr11_34621314_34621465 | 0.12 |
EHF |
ets homologous factor |
21269 |
0.24 |
| chr6_12483862_12484271 | 0.12 |
ENSG00000223321 |
. |
77053 |
0.12 |
| chr17_27054946_27055618 | 0.12 |
TLCD1 |
TLC domain containing 1 |
329 |
0.46 |
| chr8_18672049_18672314 | 0.12 |
ENSG00000251963 |
. |
5045 |
0.25 |
| chr8_61190856_61191007 | 0.12 |
CA8 |
carbonic anhydrase VIII |
3040 |
0.41 |
| chr9_124060682_124061153 | 0.12 |
GSN |
gelsolin |
1154 |
0.42 |
| chrX_38660129_38660609 | 0.11 |
MID1IP1 |
MID1 interacting protein 1 |
316 |
0.91 |
| chr4_114394825_114395109 | 0.11 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
40031 |
0.16 |
| chr15_32887434_32887644 | 0.11 |
GOLGA8N |
golgin A8 family, member N |
910 |
0.47 |
| chr4_171011358_171011655 | 0.11 |
AADAT |
aminoadipate aminotransferase |
32 |
0.99 |
| chr8_26124874_26125215 | 0.11 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
23963 |
0.24 |
| chr17_9195934_9196347 | 0.11 |
RP11-85B7.4 |
|
9649 |
0.26 |
| chr15_47797301_47797452 | 0.11 |
RP11-142J21.2 |
|
19990 |
0.2 |
| chr13_30452401_30452552 | 0.11 |
UBL3 |
ubiquitin-like 3 |
27655 |
0.23 |
| chr12_54652947_54653855 | 0.11 |
CBX5 |
chromobox homolog 5 |
25 |
0.94 |
| chr1_25654478_25654629 | 0.11 |
TMEM50A |
transmembrane protein 50A |
9855 |
0.13 |
| chr8_26497946_26498133 | 0.11 |
DPYSL2 |
dihydropyrimidinase-like 2 |
62118 |
0.14 |
| chr15_30704476_30704686 | 0.11 |
GOLGA8R |
golgin A8 family, member R |
1784 |
0.21 |
| chr1_25689807_25689958 | 0.11 |
ENSG00000238889 |
. |
17014 |
0.12 |
| chr21_15400693_15400971 | 0.11 |
AP001347.6 |
|
1060 |
0.52 |
| chr15_93572424_93572985 | 0.11 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
20608 |
0.22 |
| chr3_4458839_4458990 | 0.11 |
SUMF1 |
sulfatase modifying factor 1 |
50015 |
0.15 |
| chr7_20203384_20203535 | 0.10 |
MACC1 |
metastasis associated in colon cancer 1 |
21242 |
0.18 |
| chr20_2187871_2188239 | 0.10 |
ENSG00000263452 |
. |
50128 |
0.14 |
| chr13_28371306_28371457 | 0.10 |
GSX1 |
GS homeobox 1 |
4601 |
0.24 |
| chr10_13816413_13816564 | 0.10 |
RP11-353M9.1 |
|
45105 |
0.11 |
| chr15_76442941_76443214 | 0.10 |
RP11-593F23.1 |
|
30750 |
0.19 |
| chr10_45461690_45461881 | 0.10 |
RASSF4 |
Ras association (RalGDS/AF-6) domain family member 4 |
3887 |
0.14 |
| chr20_61196938_61197089 | 0.10 |
ENSG00000207764 |
. |
34894 |
0.1 |
| chr15_30846007_30846178 | 0.10 |
GOLGA8Q |
golgin A8 family, member Q |
1811 |
0.19 |
| chr16_85006545_85006696 | 0.10 |
ZDHHC7 |
zinc finger, DHHC-type containing 7 |
9103 |
0.23 |
| chr2_218777080_218777231 | 0.10 |
TNS1 |
tensin 1 |
6971 |
0.26 |
| chr1_157981064_157981318 | 0.10 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
14149 |
0.2 |
| chr3_64206079_64206230 | 0.10 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
4977 |
0.24 |
| chr15_32745850_32746036 | 0.10 |
GOLGA8O |
golgin A8 family, member O |
1160 |
0.32 |
| chr17_71301605_71301911 | 0.10 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
5789 |
0.21 |
| chr1_156053555_156053706 | 0.10 |
LMNA |
lamin A/C |
1261 |
0.25 |
| chr14_103465897_103466048 | 0.10 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
31103 |
0.15 |
| chr20_954428_954797 | 0.09 |
RSPO4 |
R-spondin 4 |
28292 |
0.19 |
| chr22_41684391_41684709 | 0.09 |
AL035681.1 |
|
1136 |
0.35 |
| chr8_23159016_23159167 | 0.09 |
R3HCC1 |
R3H domain and coiled-coil containing 1 |
11165 |
0.15 |
| chr1_218525548_218525699 | 0.09 |
TGFB2 |
transforming growth factor, beta 2 |
6046 |
0.23 |
| chr5_167570316_167570766 | 0.09 |
CTB-178M22.1 |
|
44183 |
0.16 |
| chr3_194979518_194980202 | 0.09 |
XXYLT1 |
xyloside xylosyltransferase 1 |
11211 |
0.17 |
| chr6_26104203_26104354 | 0.09 |
HIST1H4C |
histone cluster 1, H4c |
174 |
0.86 |
| chr15_74372869_74373020 | 0.09 |
GOLGA6A |
golgin A6 family, member A |
1947 |
0.25 |
| chr11_68081336_68081924 | 0.09 |
LRP5 |
low density lipoprotein receptor-related protein 5 |
1553 |
0.44 |
| chr21_39760697_39760848 | 0.09 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
91924 |
0.09 |
| chr5_138862247_138862421 | 0.09 |
TMEM173 |
transmembrane protein 173 |
41 |
0.97 |
| chr14_29254769_29255066 | 0.09 |
RP11-966I7.2 |
|
76 |
0.97 |
| chr15_40069729_40069880 | 0.09 |
RP11-37C7.3 |
|
4968 |
0.19 |
| chr9_18542889_18543040 | 0.09 |
ENSG00000264638 |
. |
30340 |
0.22 |
| chr10_73032227_73032416 | 0.09 |
SLC29A3 |
solute carrier family 29 (equilibrative nucleoside transporter), member 3 |
46694 |
0.12 |
| chr7_98706412_98706591 | 0.09 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
35141 |
0.17 |
| chr10_43249528_43250125 | 0.09 |
BMS1 |
BMS1 ribosome biogenesis factor |
28423 |
0.19 |
| chr1_221249282_221249543 | 0.09 |
HLX |
H2.0-like homeobox |
194828 |
0.03 |
| chr20_36652091_36652498 | 0.09 |
TTI1 |
TELO2 interacting protein 1 |
9569 |
0.16 |
| chr17_76879309_76879518 | 0.09 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
9181 |
0.14 |
| chr9_124048116_124048398 | 0.09 |
GSN-AS1 |
GSN antisense RNA 1 |
449 |
0.54 |
| chr14_95066944_95067095 | 0.09 |
SERPINA3 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
11695 |
0.16 |
| chr1_157971356_157971700 | 0.09 |
KIRREL |
kin of IRRE like (Drosophila) |
8093 |
0.22 |
| chr12_66050940_66051359 | 0.09 |
HMGA2 |
high mobility group AT-hook 2 |
166762 |
0.03 |
| chr11_11285330_11285481 | 0.09 |
RP11-567I13.1 |
|
88568 |
0.09 |
| chr2_201725693_201726476 | 0.09 |
ENSG00000252759 |
. |
1886 |
0.16 |
| chr6_151560755_151560906 | 0.08 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
304 |
0.87 |
| chr3_99379303_99379454 | 0.08 |
COL8A1 |
collagen, type VIII, alpha 1 |
21924 |
0.22 |
| chr3_12880764_12880915 | 0.08 |
ENSG00000207496 |
. |
1110 |
0.34 |
| chr13_51191217_51191680 | 0.08 |
DLEU7-AS1 |
DLEU7 antisense RNA 1 |
190544 |
0.03 |
| chr11_22647590_22648323 | 0.08 |
FANCF |
Fanconi anemia, complementation group F |
569 |
0.47 |
| chr18_77585857_77586008 | 0.08 |
KCNG2 |
potassium voltage-gated channel, subfamily G, member 2 |
37736 |
0.18 |
| chr18_43335861_43336012 | 0.08 |
RP11-116O18.3 |
|
8756 |
0.2 |
| chr16_88988440_88988637 | 0.08 |
RP11-830F9.7 |
|
15626 |
0.11 |
| chr11_77594840_77594991 | 0.08 |
RP11-91P24.7 |
|
11607 |
0.13 |
| chr5_169144327_169144497 | 0.08 |
DOCK2 |
dedicator of cytokinesis 2 |
13459 |
0.26 |
| chr12_113675872_113676102 | 0.08 |
TPCN1 |
two pore segment channel 1 |
6079 |
0.14 |
| chr16_8972208_8972493 | 0.08 |
RP11-77H9.6 |
|
9242 |
0.13 |
| chr12_27788363_27788514 | 0.08 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
25429 |
0.17 |
| chr12_46946935_46947192 | 0.08 |
SLC38A2 |
solute carrier family 38, member 2 |
180413 |
0.03 |
| chr4_87857439_87857983 | 0.08 |
AFF1 |
AF4/FMR2 family, member 1 |
149 |
0.95 |
| chr16_8716083_8716935 | 0.08 |
METTL22 |
methyltransferase like 22 |
923 |
0.61 |
| chr3_184322391_184322660 | 0.08 |
EPHB3 |
EPH receptor B3 |
42953 |
0.14 |
| chr4_10095359_10096185 | 0.08 |
ENSG00000264931 |
. |
15456 |
0.14 |
| chr1_245970690_245971117 | 0.08 |
RP11-522M21.3 |
|
131123 |
0.05 |
| chr12_125351603_125351754 | 0.08 |
SCARB1 |
scavenger receptor class B, member 1 |
3215 |
0.25 |
| chr7_129650015_129650166 | 0.08 |
RP11-306G20.1 |
|
1396 |
0.32 |
| chrX_9875291_9875592 | 0.08 |
SHROOM2 |
shroom family member 2 |
4971 |
0.24 |
| chr1_180499806_180499957 | 0.08 |
ENSG00000206905 |
. |
11626 |
0.18 |
| chr15_72948913_72949064 | 0.08 |
GOLGA6B |
golgin A6 family, member B |
1909 |
0.26 |
| chrX_67919809_67920109 | 0.08 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
6466 |
0.32 |
| chr6_158181418_158181766 | 0.08 |
SNX9 |
sorting nexin 9 |
62704 |
0.11 |
| chr15_40660789_40661084 | 0.08 |
RP11-64K12.4 |
|
905 |
0.29 |
| chr11_33795307_33795458 | 0.08 |
FBXO3 |
F-box protein 3 |
511 |
0.57 |
| chr9_132382150_132382468 | 0.07 |
C9orf50 |
chromosome 9 open reading frame 50 |
746 |
0.53 |
| chr1_6333311_6333462 | 0.07 |
GPR153 |
G protein-coupled receptor 153 |
12351 |
0.13 |
| chr17_32316789_32317089 | 0.07 |
RP11-17M24.2 |
|
13389 |
0.21 |
| chr1_35920565_35920716 | 0.07 |
KIAA0319L |
KIAA0319-like |
332 |
0.89 |
| chr3_194294141_194294292 | 0.07 |
TMEM44-AS1 |
TMEM44 antisense RNA 1 |
10524 |
0.15 |
| chr7_19151934_19152085 | 0.07 |
AC003986.6 |
|
88 |
0.96 |
| chr3_108973459_108973610 | 0.07 |
ENSG00000252889 |
. |
29558 |
0.2 |
| chr7_51098306_51098457 | 0.07 |
RP4-724E13.2 |
|
13221 |
0.29 |
| chr3_4783284_4783867 | 0.07 |
ENSG00000239126 |
. |
136811 |
0.05 |
| chr6_152411668_152411819 | 0.07 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
77743 |
0.11 |
| chr7_93698470_93698621 | 0.07 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
64851 |
0.12 |
| chr4_86954278_86954588 | 0.07 |
RP13-514E23.1 |
|
19406 |
0.19 |
| chr7_72722825_72722976 | 0.07 |
NSUN5 |
NOP2/Sun domain family, member 5 |
36 |
0.97 |
| chr7_81400181_81400399 | 0.07 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
536 |
0.87 |
| chr22_25799665_25800449 | 0.07 |
LRP5L |
low density lipoprotein receptor-related protein 5-like |
1287 |
0.46 |
| chr2_190647077_190647900 | 0.07 |
ORMDL1 |
ORM1-like 1 (S. cerevisiae) |
620 |
0.54 |
| chr2_192135580_192135731 | 0.07 |
MYO1B |
myosin IB |
5956 |
0.29 |
| chr1_39249709_39249860 | 0.07 |
RRAGC |
Ras-related GTP binding C |
75711 |
0.08 |
| chr12_27736170_27736321 | 0.07 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
16036 |
0.22 |
| chr2_20833877_20834028 | 0.07 |
HS1BP3 |
HCLS1 binding protein 3 |
4323 |
0.25 |
| chr2_74804630_74805005 | 0.07 |
LOXL3 |
lysyl oxidase-like 3 |
22000 |
0.09 |
| chr11_114002319_114002470 | 0.07 |
ENSG00000221112 |
. |
44743 |
0.15 |
| chr3_134085363_134085514 | 0.07 |
AMOTL2 |
angiomotin like 2 |
5316 |
0.24 |
| chr5_32275307_32275819 | 0.07 |
ENSG00000207052 |
. |
34205 |
0.14 |
| chr12_118745092_118745810 | 0.07 |
TAOK3 |
TAO kinase 3 |
51459 |
0.15 |
| chr4_123719852_123720003 | 0.07 |
FGF2 |
fibroblast growth factor 2 (basic) |
27936 |
0.16 |
| chr12_1062501_1062652 | 0.07 |
RAD52 |
RAD52 homolog (S. cerevisiae) |
3688 |
0.23 |
| chr3_137893746_137893897 | 0.07 |
DBR1 |
debranching RNA lariats 1 |
30 |
0.98 |
| chr18_46260220_46260632 | 0.07 |
RP11-426J5.2 |
|
45636 |
0.16 |
| chr17_2041734_2041939 | 0.07 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
34636 |
0.09 |
| chr17_56020471_56020670 | 0.06 |
CUEDC1 |
CUE domain containing 1 |
12048 |
0.16 |
| chr11_128098982_128099155 | 0.06 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
276221 |
0.01 |
| chr2_109891880_109892163 | 0.06 |
ENSG00000265965 |
. |
38060 |
0.21 |
| chr1_1309392_1309761 | 0.06 |
AURKAIP1 |
aurora kinase A interacting protein 1 |
961 |
0.28 |
| chr1_23428529_23428932 | 0.06 |
LUZP1 |
leucine zipper protein 1 |
6893 |
0.14 |
| chr4_169543840_169543991 | 0.06 |
PALLD |
palladin, cytoskeletal associated protein |
8851 |
0.22 |
| chr16_12556722_12556873 | 0.06 |
RP11-552C15.1 |
|
82542 |
0.07 |
| chr4_169402272_169402423 | 0.06 |
DDX60L |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
682 |
0.7 |
| chr11_61552248_61552399 | 0.06 |
MYRF |
myelin regulatory factor |
7245 |
0.1 |
| chr13_67501107_67501258 | 0.06 |
PCDH9-AS3 |
PCDH9 antisense RNA 3 |
50382 |
0.16 |
| chr1_10487928_10488079 | 0.06 |
APITD1 |
apoptosis-inducing, TAF9-like domain 1 |
2156 |
0.16 |
| chr2_208890523_208890891 | 0.06 |
PLEKHM3 |
pleckstrin homology domain containing, family M, member 3 |
423 |
0.8 |
| chr5_132142983_132143134 | 0.06 |
SEPT8 |
septin 8 |
125 |
0.94 |
| chr9_22165561_22165745 | 0.06 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
51976 |
0.16 |
| chr18_43409404_43409636 | 0.06 |
SIGLEC15 |
sialic acid binding Ig-like lectin 15 |
464 |
0.84 |
| chr2_166577475_166577626 | 0.06 |
GALNT3 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
40867 |
0.19 |
| chr4_173708241_173708420 | 0.06 |
ENSG00000241652 |
. |
354464 |
0.01 |
| chr10_73740928_73741079 | 0.06 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
16880 |
0.23 |
| chr8_59636475_59636734 | 0.06 |
ENSG00000238433 |
. |
61701 |
0.12 |
| chr3_127540135_127540286 | 0.06 |
MGLL |
monoglyceride lipase |
776 |
0.73 |
| chr6_133037043_133037265 | 0.06 |
VNN1 |
vanin 1 |
1966 |
0.26 |
| chr18_61088442_61089083 | 0.06 |
VPS4B |
vacuolar protein sorting 4 homolog B (S. cerevisiae) |
861 |
0.6 |
| chr5_3862538_3862689 | 0.06 |
CTD-2012M11.3 |
|
262311 |
0.02 |
| chr12_106475754_106475905 | 0.06 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
1982 |
0.4 |
| chr9_101595464_101595615 | 0.06 |
RP11-92C4.3 |
|
16356 |
0.18 |
| chr15_66906180_66906636 | 0.06 |
RP11-321F6.1 |
HCG2003567; Uncharacterized protein |
31880 |
0.13 |
| chr2_69469610_69469761 | 0.06 |
ENSG00000199460 |
. |
59676 |
0.11 |
| chr1_60746981_60747184 | 0.06 |
C1orf87 |
chromosome 1 open reading frame 87 |
207640 |
0.03 |
| chr7_27186881_27187032 | 0.06 |
HOXA-AS3 |
HOXA cluster antisense RNA 3 |
146 |
0.73 |
| chr2_9132114_9132265 | 0.06 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
11605 |
0.23 |
| chr5_172304861_172305060 | 0.06 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
27260 |
0.16 |
| chr2_172958775_172959188 | 0.06 |
DLX2 |
distal-less homeobox 2 |
8647 |
0.2 |
| chr21_18885134_18885388 | 0.06 |
CXADR |
coxsackie virus and adenovirus receptor |
103 |
0.96 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0055026 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.0 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |