Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for ETV6
Z-value: 1.73
Transcription factors associated with ETV6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV6
|
ENSG00000139083.6 | ETS variant transcription factor 6 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_11993430_11993581 | ETV6 | 45366 | 0.189977 | 0.90 | 1.0e-03 | Click! |
| chr12_11954306_11954457 | ETV6 | 48946 | 0.178414 | 0.88 | 1.9e-03 | Click! |
| chr12_11971660_11971811 | ETV6 | 66300 | 0.133982 | 0.83 | 5.3e-03 | Click! |
| chr12_11819592_11819743 | ETV6 | 16879 | 0.256524 | 0.83 | 5.9e-03 | Click! |
| chr12_11819415_11819566 | ETV6 | 16702 | 0.256924 | 0.82 | 6.3e-03 | Click! |
Activity of the ETV6 motif across conditions
Conditions sorted by the z-value of the ETV6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
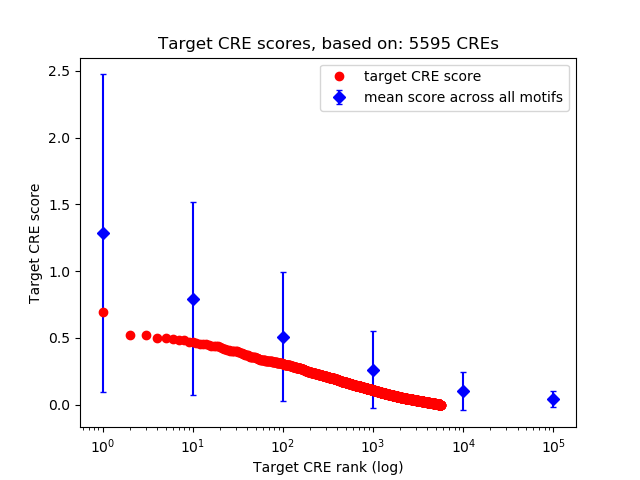
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_134162105_134162300 | 0.69 |
TG |
thyroglobulin |
36455 |
0.15 |
| chr16_53127962_53128318 | 0.52 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
4932 |
0.27 |
| chr4_185269211_185269362 | 0.52 |
ENSG00000244512 |
. |
69197 |
0.09 |
| chr11_72498251_72498402 | 0.50 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
1314 |
0.31 |
| chr11_59495823_59496091 | 0.50 |
OR10V1 |
olfactory receptor, family 10, subfamily V, member 1 |
14620 |
0.12 |
| chr21_40382908_40383154 | 0.49 |
ENSG00000272015 |
. |
116322 |
0.06 |
| chr19_6887176_6887376 | 0.49 |
EMR1 |
egf-like module containing, mucin-like, hormone receptor-like 1 |
301 |
0.88 |
| chr6_44201103_44201369 | 0.48 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
6360 |
0.12 |
| chr5_39228625_39228776 | 0.47 |
FYB |
FYN binding protein |
8992 |
0.29 |
| chr1_21863061_21863212 | 0.47 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
14636 |
0.19 |
| chr8_130696046_130696267 | 0.46 |
GSDMC |
gasdermin C |
102978 |
0.06 |
| chr3_194456653_194456804 | 0.46 |
FAM43A |
family with sequence similarity 43, member A |
50106 |
0.11 |
| chr6_145099214_145099365 | 0.46 |
UTRN |
utrophin |
11076 |
0.33 |
| chr2_99872403_99872554 | 0.45 |
LYG2 |
lysozyme G-like 2 |
733 |
0.65 |
| chr14_89950953_89951233 | 0.45 |
FOXN3 |
forkhead box N3 |
9302 |
0.21 |
| chr21_34439817_34440107 | 0.44 |
OLIG1 |
oligodendrocyte transcription factor 1 |
2488 |
0.25 |
| chr1_47052866_47053017 | 0.44 |
MKNK1 |
MAP kinase interacting serine/threonine kinase 1 |
1010 |
0.47 |
| chr4_86618813_86618964 | 0.44 |
ENSG00000266421 |
. |
24733 |
0.25 |
| chr3_60833462_60833730 | 0.44 |
ENSG00000212211 |
. |
8681 |
0.34 |
| chr13_49790716_49790984 | 0.43 |
MLNR |
motilin receptor |
3624 |
0.28 |
| chr3_14459825_14460205 | 0.42 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
14163 |
0.22 |
| chr13_32621986_32622137 | 0.42 |
FRY |
furry homolog (Drosophila) |
12940 |
0.22 |
| chr5_173145182_173145333 | 0.41 |
ENSG00000263401 |
. |
11824 |
0.27 |
| chr7_115980158_115980451 | 0.41 |
ENSG00000216076 |
. |
5694 |
0.22 |
| chr8_37761152_37761429 | 0.41 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
4305 |
0.16 |
| chr21_37558737_37558888 | 0.41 |
DOPEY2 |
dopey family member 2 |
21979 |
0.12 |
| chr1_167127932_167128119 | 0.41 |
RP11-277B15.2 |
|
36692 |
0.13 |
| chr2_10538806_10538994 | 0.40 |
HPCAL1 |
hippocalcin-like 1 |
21247 |
0.18 |
| chr11_59823736_59824267 | 0.40 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
59 |
0.97 |
| chr14_21025165_21025316 | 0.40 |
RNASE9 |
ribonuclease, RNase A family, 9 (non-active) |
3850 |
0.14 |
| chr3_39323163_39323419 | 0.40 |
CX3CR1 |
chemokine (C-X3-C motif) receptor 1 |
65 |
0.98 |
| chr6_6724343_6724679 | 0.39 |
LY86-AS1 |
LY86 antisense RNA 1 |
101507 |
0.08 |
| chr17_1674840_1675255 | 0.39 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
34 |
0.96 |
| chr11_78062676_78062877 | 0.39 |
GAB2 |
GRB2-associated binding protein 2 |
9850 |
0.18 |
| chr4_40623166_40623743 | 0.39 |
RBM47 |
RNA binding motif protein 47 |
8427 |
0.27 |
| chr9_93567526_93567713 | 0.38 |
SYK |
spleen tyrosine kinase |
3410 |
0.4 |
| chr6_12963445_12963682 | 0.38 |
PHACTR1 |
phosphatase and actin regulator 1 |
5349 |
0.34 |
| chr5_175101372_175101562 | 0.37 |
HRH2 |
histamine receptor H2 |
6997 |
0.22 |
| chr4_26220381_26220779 | 0.37 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
53649 |
0.18 |
| chr15_69990444_69990636 | 0.37 |
ENSG00000238870 |
. |
32621 |
0.21 |
| chr3_72572531_72572682 | 0.37 |
ENSG00000238568 |
. |
49510 |
0.14 |
| chr20_39123475_39123626 | 0.36 |
ENSG00000252434 |
. |
78035 |
0.12 |
| chr8_48309430_48309581 | 0.36 |
SPIDR |
scaffolding protein involved in DNA repair |
43458 |
0.19 |
| chr1_25304935_25305322 | 0.36 |
RUNX3 |
runt-related transcription factor 3 |
13627 |
0.19 |
| chr10_73396625_73396776 | 0.36 |
CDH23 |
cadherin-related 23 |
8973 |
0.26 |
| chr18_77403247_77403434 | 0.36 |
CTDP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
36461 |
0.19 |
| chr20_44538606_44538964 | 0.36 |
PLTP |
phospholipid transfer protein |
791 |
0.42 |
| chr17_27346283_27346434 | 0.36 |
AC024619.2 |
|
3998 |
0.13 |
| chr8_141123630_141123910 | 0.36 |
C8orf17 |
chromosome 8 open reading frame 17 |
180354 |
0.03 |
| chr19_2188091_2188301 | 0.35 |
DOT1L |
DOT1-like histone H3K79 methyltransferase |
24012 |
0.08 |
| chr1_42195372_42195655 | 0.35 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
28841 |
0.2 |
| chr19_6672050_6672201 | 0.35 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
1526 |
0.31 |
| chr8_8966758_8966909 | 0.35 |
RP11-10A14.3 |
|
32101 |
0.12 |
| chr10_26626064_26626215 | 0.35 |
APBB1IP |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
100993 |
0.08 |
| chr18_77269072_77269296 | 0.34 |
AC018445.1 |
Uncharacterized protein |
6873 |
0.29 |
| chr2_113597888_113598039 | 0.34 |
IL1B |
interleukin 1, beta |
3483 |
0.2 |
| chr13_32685646_32685859 | 0.34 |
FRY |
furry homolog (Drosophila) |
50751 |
0.16 |
| chr1_161211637_161212023 | 0.33 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
3738 |
0.09 |
| chrX_108977400_108977585 | 0.33 |
ACSL4 |
acyl-CoA synthetase long-chain family member 4 |
860 |
0.67 |
| chrX_25067386_25067645 | 0.33 |
ENSG00000240439 |
. |
11541 |
0.25 |
| chr1_32123678_32123829 | 0.33 |
RP11-73M7.6 |
|
894 |
0.46 |
| chr3_184095207_184095533 | 0.33 |
THPO |
thrombopoietin |
558 |
0.59 |
| chr12_10204798_10205132 | 0.33 |
CLEC9A |
C-type lectin domain family 9, member A |
21689 |
0.1 |
| chr15_70005461_70005680 | 0.33 |
ENSG00000238870 |
. |
17591 |
0.25 |
| chr15_67158530_67158864 | 0.33 |
SMAD6 |
SMAD family member 6 |
154662 |
0.04 |
| chr20_1878767_1878918 | 0.33 |
SIRPA |
signal-regulatory protein alpha |
2900 |
0.32 |
| chr4_8270805_8271133 | 0.33 |
HTRA3 |
HtrA serine peptidase 3 |
523 |
0.82 |
| chr1_36952288_36952439 | 0.33 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
3484 |
0.21 |
| chr6_37984799_37985021 | 0.33 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
87175 |
0.09 |
| chr2_26549164_26549440 | 0.33 |
GPR113 |
G protein-coupled receptor 113 |
7332 |
0.2 |
| chr6_146917295_146917531 | 0.33 |
ADGB |
androglobin |
2688 |
0.34 |
| chr1_33514881_33515110 | 0.33 |
AK2 |
adenylate kinase 2 |
12402 |
0.17 |
| chr10_48483820_48483971 | 0.33 |
GDF10 |
growth differentiation factor 10 |
44919 |
0.14 |
| chr19_49920997_49921157 | 0.32 |
PTH2 |
parathyroid hormone 2 |
5621 |
0.07 |
| chr14_70111444_70111741 | 0.32 |
KIAA0247 |
KIAA0247 |
33279 |
0.18 |
| chr4_2806746_2806916 | 0.32 |
SH3BP2 |
SH3-domain binding protein 2 |
6106 |
0.21 |
| chr7_2832108_2832259 | 0.32 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
22709 |
0.22 |
| chr11_3845898_3846068 | 0.32 |
RHOG |
ras homolog family member G |
5898 |
0.1 |
| chr19_52074425_52074576 | 0.32 |
ZNF175 |
zinc finger protein 175 |
51 |
0.96 |
| chr14_65509694_65510092 | 0.32 |
ENSG00000266531 |
. |
1513 |
0.32 |
| chr1_22454794_22454972 | 0.32 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
1461 |
0.43 |
| chr3_47338789_47338970 | 0.32 |
KIF9 |
kinesin family member 9 |
13938 |
0.16 |
| chr2_57827496_57827758 | 0.32 |
ENSG00000212168 |
. |
55957 |
0.18 |
| chr8_61798114_61798398 | 0.32 |
RP11-33I11.2 |
|
76091 |
0.12 |
| chrX_123517274_123517517 | 0.32 |
SH2D1A |
SH2 domain containing 1A |
36956 |
0.23 |
| chr10_94547379_94547538 | 0.32 |
EXOC6 |
exocyst complex component 6 |
43477 |
0.16 |
| chr2_43460695_43460846 | 0.32 |
AC010883.5 |
|
4058 |
0.27 |
| chr3_17076946_17077097 | 0.31 |
PLCL2-AS1 |
PLCL2 antisense RNA 1 |
8663 |
0.24 |
| chr7_74498503_74498654 | 0.31 |
WBSCR16 |
Williams-Beuren syndrome chromosome region 16 |
8514 |
0.22 |
| chr20_1800907_1801169 | 0.31 |
SIRPA |
signal-regulatory protein alpha |
74116 |
0.1 |
| chr5_135347652_135347946 | 0.31 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
16785 |
0.2 |
| chr12_56371376_56371555 | 0.31 |
RAB5B |
RAB5B, member RAS oncogene family |
3580 |
0.1 |
| chr17_56361536_56361794 | 0.31 |
MPO |
myeloperoxidase |
3369 |
0.16 |
| chr19_14898095_14898371 | 0.31 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
8880 |
0.14 |
| chr4_185924099_185924250 | 0.31 |
HELT |
helt bHLH transcription factor |
15821 |
0.22 |
| chr16_66577982_66578230 | 0.31 |
TK2 |
thymidine kinase 2, mitochondrial |
5595 |
0.1 |
| chr18_24356811_24356962 | 0.31 |
AQP4 |
aquaporin 4 |
85926 |
0.08 |
| chr8_29242547_29242792 | 0.31 |
RP4-676L2.1 |
|
31982 |
0.15 |
| chr7_141875353_141875504 | 0.31 |
RP11-1220K2.2 |
Putative inactive maltase-glucoamylase-like protein LOC93432 |
63874 |
0.1 |
| chr20_58713423_58713574 | 0.31 |
C20orf197 |
chromosome 20 open reading frame 197 |
82518 |
0.1 |
| chr8_61851808_61852029 | 0.31 |
CLVS1 |
clavesin 1 |
117799 |
0.06 |
| chr17_74099339_74099490 | 0.31 |
EXOC7 |
exocyst complex component 7 |
63 |
0.95 |
| chr4_152774376_152774527 | 0.30 |
PET112 |
PET112 homolog (yeast) |
92276 |
0.08 |
| chr2_65296545_65296800 | 0.30 |
CEP68 |
centrosomal protein 68kDa |
13073 |
0.17 |
| chr14_75714237_75714434 | 0.30 |
RP11-293M10.2 |
|
11779 |
0.15 |
| chr2_109872100_109872486 | 0.30 |
ENSG00000265965 |
. |
57788 |
0.15 |
| chr19_33771380_33771704 | 0.30 |
CTD-2540B15.11 |
|
19298 |
0.12 |
| chr6_139090481_139090632 | 0.30 |
CCDC28A |
coiled-coil domain containing 28A |
4101 |
0.24 |
| chr15_77713368_77713519 | 0.30 |
HMG20A |
high mobility group 20A |
90 |
0.95 |
| chr3_113326958_113327109 | 0.30 |
ENSG00000265253 |
. |
13310 |
0.15 |
| chr16_58889405_58889622 | 0.30 |
ENSG00000244003 |
. |
85085 |
0.09 |
| chr16_84377709_84377860 | 0.30 |
ATP2C2 |
ATPase, Ca++ transporting, type 2C, member 2 |
24349 |
0.15 |
| chr5_139594871_139595091 | 0.29 |
CTB-131B5.2 |
|
15233 |
0.14 |
| chr10_126696899_126697050 | 0.29 |
CTBP2 |
C-terminal binding protein 2 |
2392 |
0.3 |
| chr15_31700478_31700629 | 0.29 |
KLF13 |
Kruppel-like factor 13 |
42196 |
0.21 |
| chr22_42832185_42832336 | 0.29 |
NFAM1 |
NFAT activating protein with ITAM motif 1 |
3859 |
0.23 |
| chr12_78393170_78393321 | 0.29 |
NAV3 |
neuron navigator 3 |
33189 |
0.24 |
| chr1_146675856_146676007 | 0.29 |
FMO5 |
flavin containing monooxygenase 5 |
20970 |
0.14 |
| chr2_64610358_64610618 | 0.29 |
ENSG00000264297 |
. |
42595 |
0.16 |
| chr5_81133416_81133809 | 0.29 |
SSBP2 |
single-stranded DNA binding protein 2 |
86540 |
0.09 |
| chr11_22829816_22830016 | 0.29 |
RP11-17A1.3 |
|
21010 |
0.18 |
| chr7_157129016_157129203 | 0.29 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
551 |
0.82 |
| chr15_78691765_78692045 | 0.29 |
IREB2 |
iron-responsive element binding protein 2 |
37868 |
0.12 |
| chr1_212768754_212768905 | 0.29 |
ATF3 |
activating transcription factor 3 |
13183 |
0.15 |
| chr4_37948267_37948441 | 0.29 |
PTTG2 |
pituitary tumor-transforming 2 |
13702 |
0.24 |
| chr1_183544894_183545045 | 0.29 |
NCF2 |
neutrophil cytosolic factor 2 |
6609 |
0.21 |
| chr3_128532158_128532319 | 0.29 |
RAB7A |
RAB7A, member RAS oncogene family |
18036 |
0.16 |
| chr17_8324143_8324299 | 0.29 |
NDEL1 |
nudE neurodevelopment protein 1-like 1 |
7772 |
0.12 |
| chr21_43431068_43431344 | 0.29 |
ZBTB21 |
zinc finger and BTB domain containing 21 |
710 |
0.7 |
| chr1_152492276_152492444 | 0.28 |
CRCT1 |
cysteine-rich C-terminal 1 |
5382 |
0.15 |
| chr3_138620560_138620746 | 0.28 |
FOXL2 |
forkhead box L2 |
45329 |
0.13 |
| chr11_65584293_65584444 | 0.28 |
SNX32 |
sorting nexin 32 |
16744 |
0.08 |
| chr4_8349838_8350141 | 0.28 |
ENSG00000202054 |
. |
44753 |
0.14 |
| chr13_29148843_29149031 | 0.28 |
FLT1 |
fms-related tyrosine kinase 1 |
79672 |
0.1 |
| chr4_86590486_86590637 | 0.28 |
ENSG00000266421 |
. |
53060 |
0.16 |
| chrX_119150159_119150382 | 0.28 |
RP4-755D9.1 |
|
19931 |
0.17 |
| chr2_119130151_119130302 | 0.28 |
INSIG2 |
insulin induced gene 2 |
284176 |
0.01 |
| chr13_60576787_60576938 | 0.28 |
DIAPH3-AS1 |
DIAPH3 antisense RNA 1 |
10023 |
0.25 |
| chr9_124299702_124300078 | 0.28 |
ENSG00000252940 |
. |
2265 |
0.29 |
| chr19_19143693_19143980 | 0.28 |
SUGP2 |
SURP and G patch domain containing 2 |
407 |
0.54 |
| chr10_134198535_134198861 | 0.28 |
PWWP2B |
PWWP domain containing 2B |
11974 |
0.18 |
| chr15_69606302_69606478 | 0.28 |
PAQR5 |
progestin and adipoQ receptor family member V |
352 |
0.82 |
| chr8_10669979_10670130 | 0.28 |
PINX1 |
PIN2/TERF1 interacting, telomerase inhibitor 1 |
27227 |
0.12 |
| chr3_151107193_151107344 | 0.28 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
4668 |
0.23 |
| chr11_61683909_61684462 | 0.28 |
RAB3IL1 |
RAB3A interacting protein (rabin3)-like 1 |
812 |
0.51 |
| chr1_175097367_175097518 | 0.27 |
TNN |
tenascin N |
60448 |
0.11 |
| chr12_27153812_27154240 | 0.27 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
2192 |
0.25 |
| chr12_32629297_32629583 | 0.27 |
ENSG00000253063 |
. |
965 |
0.61 |
| chr9_132316854_132317005 | 0.27 |
RP11-492E3.2 |
|
20759 |
0.14 |
| chr11_113968901_113969117 | 0.27 |
ENSG00000221112 |
. |
11358 |
0.23 |
| chr22_30648620_30649241 | 0.27 |
LIF |
leukemia inhibitory factor |
6090 |
0.11 |
| chr11_43855946_43856097 | 0.27 |
RP11-613D13.5 |
|
529 |
0.81 |
| chr14_59827605_59828209 | 0.27 |
ENSG00000252869 |
. |
38834 |
0.18 |
| chr15_65139770_65140076 | 0.27 |
AC069368.3 |
Uncharacterized protein |
5776 |
0.14 |
| chr6_15428120_15428372 | 0.27 |
JARID2 |
jumonji, AT rich interactive domain 2 |
27157 |
0.23 |
| chr16_84766620_84766890 | 0.27 |
USP10 |
ubiquitin specific peptidase 10 |
33109 |
0.16 |
| chr6_116889940_116890146 | 0.27 |
RWDD1 |
RWD domain containing 1 |
2487 |
0.21 |
| chr14_107081781_107081932 | 0.27 |
IGHV4-59 |
immunoglobulin heavy variable 4-59 |
1869 |
0.07 |
| chr10_105785288_105785439 | 0.27 |
COL17A1 |
collagen, type XVII, alpha 1 |
7363 |
0.18 |
| chr16_31141073_31141404 | 0.27 |
RP11-388M20.2 |
|
1516 |
0.15 |
| chr1_109742432_109742583 | 0.27 |
ENSG00000238310 |
. |
7963 |
0.15 |
| chr2_73207068_73207254 | 0.27 |
EMX1 |
empty spiracles homeobox 1 |
56044 |
0.12 |
| chr16_81498895_81499086 | 0.27 |
CMIP |
c-Maf inducing protein |
20215 |
0.24 |
| chr6_126307498_126307649 | 0.27 |
TRMT11 |
tRNA methyltransferase 11 homolog (S. cerevisiae) |
3 |
0.98 |
| chr1_208017036_208017239 | 0.27 |
ENSG00000203709 |
. |
41269 |
0.16 |
| chr7_47846848_47846999 | 0.27 |
PKD1L1 |
polycystic kidney disease 1 like 1 |
5912 |
0.22 |
| chr8_116470203_116470515 | 0.27 |
TRPS1 |
trichorhinophalangeal syndrome I |
34089 |
0.22 |
| chr2_24292097_24292248 | 0.27 |
SF3B14 |
Pre-mRNA branch site protein p14 |
7141 |
0.11 |
| chr2_86102638_86102927 | 0.26 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
8005 |
0.21 |
| chr9_125059861_125060215 | 0.26 |
ENSG00000264227 |
. |
1604 |
0.3 |
| chr20_33872479_33872715 | 0.26 |
EIF6 |
eukaryotic translation initiation factor 6 |
24 |
0.87 |
| chr2_149303357_149303735 | 0.26 |
MBD5 |
methyl-CpG binding domain protein 5 |
77252 |
0.11 |
| chr19_42051966_42052728 | 0.26 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
3539 |
0.21 |
| chr7_100400756_100400965 | 0.26 |
ENSG00000263672 |
. |
18026 |
0.1 |
| chr7_138720207_138720441 | 0.26 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
451 |
0.85 |
| chr14_60619421_60619786 | 0.26 |
DHRS7 |
dehydrogenase/reductase (SDR family) member 7 |
289 |
0.92 |
| chr17_44983331_44983711 | 0.26 |
ENSG00000210709 |
. |
14969 |
0.15 |
| chr2_183989474_183989788 | 0.26 |
NUP35 |
nucleoporin 35kDa |
458 |
0.82 |
| chr5_177667186_177667397 | 0.26 |
PHYKPL |
5-phosphohydroxy-L-lysine phospho-lyase |
7505 |
0.21 |
| chr8_67693713_67693929 | 0.26 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
6020 |
0.24 |
| chr10_64612517_64612668 | 0.26 |
EGR2 |
early growth response 2 |
33665 |
0.17 |
| chr15_43562661_43562823 | 0.25 |
TGM5 |
transglutaminase 5 |
3687 |
0.15 |
| chr7_127693207_127693470 | 0.25 |
LRRC4 |
leucine rich repeat containing 4 |
21178 |
0.2 |
| chr16_58058882_58059051 | 0.25 |
MMP15 |
matrix metallopeptidase 15 (membrane-inserted) |
504 |
0.72 |
| chr22_38023733_38023903 | 0.25 |
SH3BP1 |
SH3-domain binding protein 1 |
11664 |
0.1 |
| chr2_190424109_190424405 | 0.25 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
21356 |
0.22 |
| chr10_51504373_51504582 | 0.25 |
AGAP7 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 7 |
18150 |
0.18 |
| chr9_123433443_123433594 | 0.25 |
MEGF9 |
multiple EGF-like-domains 9 |
43094 |
0.17 |
| chr9_113092238_113092389 | 0.25 |
TXNDC8 |
thioredoxin domain containing 8 (spermatozoa) |
7775 |
0.23 |
| chr12_54686387_54686538 | 0.25 |
NFE2 |
nuclear factor, erythroid 2 |
3080 |
0.11 |
| chr15_75761353_75761612 | 0.25 |
SIN3A |
SIN3 transcription regulator family member A |
13299 |
0.17 |
| chr7_38295749_38295900 | 0.25 |
STARD3NL |
STARD3 N-terminal like |
77827 |
0.12 |
| chr12_49360293_49360446 | 0.25 |
WNT10B |
wingless-type MMTV integration site family, member 10B |
4170 |
0.08 |
| chr4_142558013_142558295 | 0.25 |
IL15 |
interleukin 15 |
54 |
0.99 |
| chr2_216371790_216371973 | 0.24 |
AC012462.1 |
|
70905 |
0.11 |
| chr17_77755803_77755954 | 0.24 |
CBX2 |
chromobox homolog 2 |
3885 |
0.16 |
| chr5_55904489_55904655 | 0.24 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
2513 |
0.36 |
| chr11_128236040_128236191 | 0.24 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
139174 |
0.05 |
| chr18_2653058_2653283 | 0.24 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
2567 |
0.22 |
| chr5_171839971_171840157 | 0.24 |
ENSG00000216127 |
. |
26694 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 0.4 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.4 | GO:0009415 | response to water(GO:0009415) |
| 0.1 | 0.2 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 0.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.2 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.2 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.1 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.3 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0032329 | serine transport(GO:0032329) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0031392 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0032725 | positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.2 | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) |
| 0.0 | 0.1 | GO:2000319 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.2 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.0 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0090594 | wound healing involved in inflammatory response(GO:0002246) connective tissue replacement involved in inflammatory response wound healing(GO:0002248) inflammatory response to wounding(GO:0090594) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.0 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:1904747 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.2 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.0 | GO:0001820 | serotonin secretion(GO:0001820) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 0.0 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0070849 | response to epidermal growth factor(GO:0070849) |
| 0.0 | 0.1 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.0 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:1990266 | neutrophil chemotaxis(GO:0030593) neutrophil migration(GO:1990266) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.0 | 0.0 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.0 | GO:0090494 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0030540 | female genitalia development(GO:0030540) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.0 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 0.0 | 0.0 | GO:0034444 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.0 | 0.0 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.0 | GO:0031935 | regulation of chromatin silencing(GO:0031935) negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.0 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0051138 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.0 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |