Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
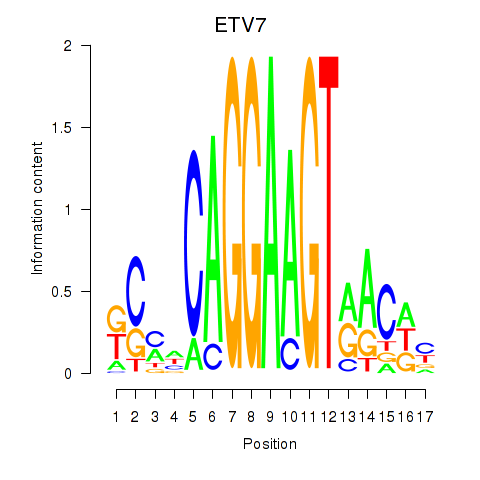
Results for ETV7
Z-value: 1.16
Transcription factors associated with ETV7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ETV7
|
ENSG00000010030.9 | ETS variant transcription factor 7 |
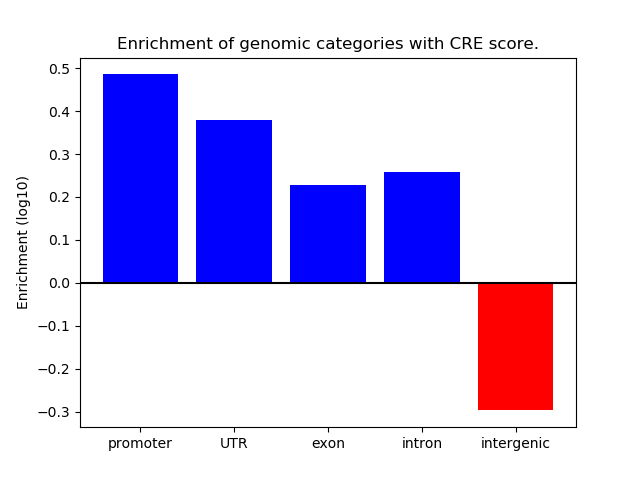
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_36355012_36355199 | ETV7 | 388 | 0.543896 | -0.33 | 3.9e-01 | Click! |
| chr6_36355753_36356314 | ETV7 | 131 | 0.933451 | 0.07 | 8.7e-01 | Click! |
| chr6_36355421_36355572 | ETV7 | 3 | 0.945827 | -0.00 | 1.0e+00 | Click! |
Activity of the ETV7 motif across conditions
Conditions sorted by the z-value of the ETV7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
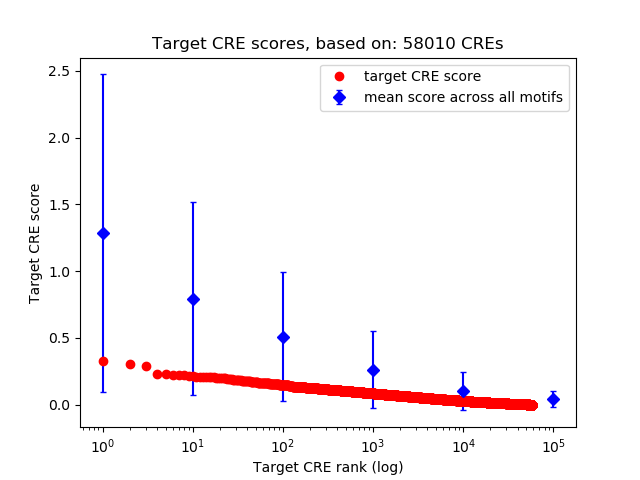
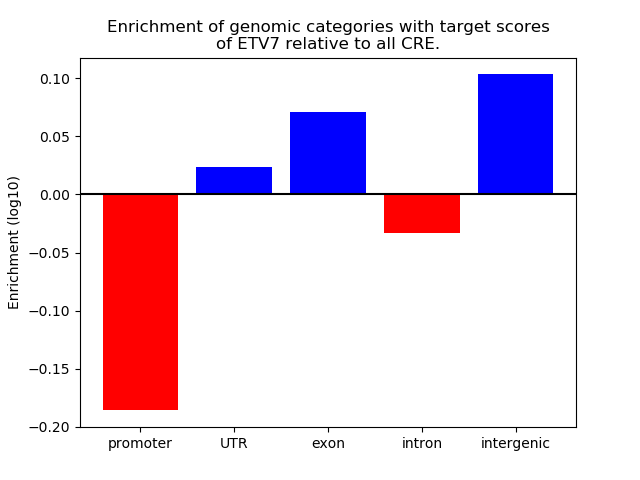
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_13697746_13698201 | 0.32 |
FAR1-IT1 |
FAR1 intronic transcript 1 (non-protein coding) |
7099 |
0.2 |
| chr14_51886452_51886750 | 0.30 |
FRMD6 |
FERM domain containing 6 |
69254 |
0.11 |
| chr4_154581482_154581754 | 0.29 |
RP11-153M7.3 |
|
17793 |
0.19 |
| chr9_137233668_137233871 | 0.23 |
RXRA |
retinoid X receptor, alpha |
15343 |
0.23 |
| chr1_92846574_92846807 | 0.23 |
ENSG00000242764 |
. |
21552 |
0.21 |
| chr7_28723084_28723448 | 0.22 |
CREB5 |
cAMP responsive element binding protein 5 |
2332 |
0.46 |
| chrX_13269027_13269605 | 0.22 |
GS1-600G8.3 |
|
59455 |
0.14 |
| chr2_7144203_7144376 | 0.22 |
RNF144A |
ring finger protein 144A |
7218 |
0.26 |
| chr17_32570710_32570941 | 0.22 |
CCL2 |
chemokine (C-C motif) ligand 2 |
11479 |
0.13 |
| chr11_35873645_35873874 | 0.21 |
ENSG00000266590 |
. |
8535 |
0.22 |
| chr6_48850860_48851011 | 0.21 |
ENSG00000221175 |
. |
33375 |
0.25 |
| chr8_142136676_142136992 | 0.21 |
RP11-809O17.1 |
|
1386 |
0.38 |
| chr2_113597075_113597265 | 0.21 |
IL1B |
interleukin 1, beta |
2690 |
0.24 |
| chr10_5644211_5644362 | 0.21 |
ENSG00000240577 |
. |
29140 |
0.14 |
| chr16_57717963_57718157 | 0.21 |
CCDC135 |
coiled-coil domain containing 135 |
10645 |
0.12 |
| chr10_60400963_60401249 | 0.20 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
128206 |
0.06 |
| chr5_150477041_150477197 | 0.20 |
TNIP1 |
TNFAIP3 interacting protein 1 |
3981 |
0.23 |
| chr6_167142675_167142845 | 0.20 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
28639 |
0.23 |
| chr9_224674_224929 | 0.20 |
C9orf66 |
chromosome 9 open reading frame 66 |
8908 |
0.18 |
| chr8_20004646_20004840 | 0.20 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
33732 |
0.16 |
| chr17_29280275_29280468 | 0.20 |
AC091177.1 |
|
3608 |
0.12 |
| chr5_177859454_177859724 | 0.20 |
CTB-26E19.1 |
|
10506 |
0.23 |
| chr5_149837604_149837792 | 0.20 |
RPS14 |
ribosomal protein S14 |
8379 |
0.18 |
| chr10_135093978_135094209 | 0.20 |
ADAM8 |
ADAM metallopeptidase domain 8 |
3721 |
0.12 |
| chr4_185819623_185819887 | 0.19 |
ENSG00000263458 |
. |
39839 |
0.14 |
| chr2_100388072_100388250 | 0.19 |
AFF3 |
AF4/FMR2 family, member 3 |
193329 |
0.03 |
| chr2_85777153_85777517 | 0.19 |
MAT2A |
methionine adenosyltransferase II, alpha |
10570 |
0.09 |
| chr11_105052468_105053090 | 0.19 |
CARD18 |
caspase recruitment domain family, member 18 |
42326 |
0.16 |
| chr2_60598504_60598911 | 0.19 |
ENSG00000200807 |
. |
13033 |
0.23 |
| chr2_118856015_118856215 | 0.19 |
INSIG2 |
insulin induced gene 2 |
10065 |
0.24 |
| chr9_92275563_92275780 | 0.19 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
55718 |
0.16 |
| chr13_45376199_45376477 | 0.18 |
ENSG00000238932 |
. |
173529 |
0.03 |
| chr18_43381728_43382214 | 0.18 |
SIGLEC15 |
sialic acid binding Ig-like lectin 15 |
23506 |
0.18 |
| chr2_166672118_166672418 | 0.18 |
ENSG00000221234 |
. |
1543 |
0.44 |
| chr11_43713235_43713438 | 0.18 |
HSD17B12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
11005 |
0.26 |
| chr3_151139810_151140059 | 0.18 |
IGSF10 |
immunoglobulin superfamily, member 10 |
21004 |
0.19 |
| chr21_40107907_40108234 | 0.18 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
69161 |
0.12 |
| chr12_93521110_93521555 | 0.18 |
RP11-511B23.2 |
|
11535 |
0.21 |
| chr8_37758588_37758772 | 0.18 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
1695 |
0.28 |
| chr15_42415254_42415667 | 0.18 |
PLA2G4D |
phospholipase A2, group IVD (cytosolic) |
28708 |
0.13 |
| chr10_98416000_98416373 | 0.18 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
13182 |
0.2 |
| chr2_75095702_75095909 | 0.18 |
HK2 |
hexokinase 2 |
33508 |
0.19 |
| chr15_90755314_90755522 | 0.17 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
9564 |
0.12 |
| chr4_154576776_154576934 | 0.17 |
RP11-153M7.3 |
|
22556 |
0.19 |
| chr1_22454794_22454972 | 0.17 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
1461 |
0.43 |
| chr3_185471276_185471520 | 0.17 |
ENSG00000265470 |
. |
14294 |
0.21 |
| chr6_90111426_90111593 | 0.17 |
RRAGD |
Ras-related GTP binding D |
10280 |
0.19 |
| chr5_134769606_134770119 | 0.17 |
C5orf20 |
chromosome 5 open reading frame 20 |
13176 |
0.14 |
| chr12_55276178_55276393 | 0.17 |
MUCL1 |
mucin-like 1 |
27964 |
0.2 |
| chr2_121363772_121364036 | 0.17 |
ENSG00000201006 |
. |
44945 |
0.19 |
| chr8_82029068_82029440 | 0.17 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
4951 |
0.32 |
| chr5_143522492_143522714 | 0.17 |
ENSG00000239390 |
. |
2159 |
0.35 |
| chr12_52099319_52099549 | 0.17 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
19227 |
0.23 |
| chr5_157661766_157661969 | 0.17 |
ENSG00000222626 |
. |
257903 |
0.02 |
| chr15_33735902_33736053 | 0.17 |
RYR3 |
ryanodine receptor 3 |
132800 |
0.05 |
| chr20_58635186_58635347 | 0.16 |
C20orf197 |
chromosome 20 open reading frame 197 |
4286 |
0.29 |
| chr8_142120178_142120491 | 0.16 |
DENND3 |
DENN/MADD domain containing 3 |
7043 |
0.22 |
| chr3_37372099_37372250 | 0.16 |
ENSG00000222208 |
. |
10966 |
0.2 |
| chr8_62644087_62644326 | 0.16 |
ENSG00000264408 |
. |
16859 |
0.22 |
| chr8_72762424_72762575 | 0.16 |
MSC |
musculin |
5796 |
0.19 |
| chr7_100411285_100411882 | 0.16 |
ENSG00000263672 |
. |
7303 |
0.11 |
| chr12_117514666_117514937 | 0.16 |
TESC |
tescalcin |
22450 |
0.23 |
| chr17_46080195_46080690 | 0.16 |
RP11-6N17.10 |
|
6880 |
0.1 |
| chr13_32822854_32823183 | 0.16 |
FRY |
furry homolog (Drosophila) |
15787 |
0.21 |
| chrX_20413169_20413368 | 0.16 |
ENSG00000252978 |
. |
56958 |
0.16 |
| chr12_11962461_11962612 | 0.16 |
ETV6 |
ets variant 6 |
57101 |
0.16 |
| chr13_81205127_81205302 | 0.16 |
ENSG00000202398 |
. |
261545 |
0.02 |
| chr8_48318969_48319120 | 0.16 |
SPIDR |
scaffolding protein involved in DNA repair |
33919 |
0.22 |
| chr4_185179575_185179726 | 0.16 |
ENSG00000221523 |
. |
8934 |
0.21 |
| chr15_42801149_42801309 | 0.16 |
SNAP23 |
synaptosomal-associated protein, 23kDa |
2816 |
0.19 |
| chr6_44202164_44202547 | 0.16 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
7479 |
0.11 |
| chr12_54688458_54688720 | 0.16 |
NFE2 |
nuclear factor, erythroid 2 |
953 |
0.32 |
| chr3_127487286_127487461 | 0.16 |
MGLL |
monoglyceride lipase |
15478 |
0.23 |
| chr7_102631323_102631632 | 0.16 |
NFE4 |
nuclear factor, erythroid 4 |
17508 |
0.14 |
| chr17_79879033_79879418 | 0.16 |
SIRT7 |
sirtuin 7 |
26 |
0.9 |
| chr11_111394946_111395097 | 0.15 |
RP11-794P6.6 |
|
2459 |
0.16 |
| chr6_6804228_6804533 | 0.15 |
ENSG00000240936 |
. |
134750 |
0.05 |
| chr14_92506646_92506800 | 0.15 |
TRIP11 |
thyroid hormone receptor interactor 11 |
320 |
0.87 |
| chr1_203254365_203254586 | 0.15 |
BTG2 |
BTG family, member 2 |
20189 |
0.15 |
| chr6_71958092_71958555 | 0.15 |
RP11-154D6.1 |
|
36312 |
0.16 |
| chr1_239979756_239979955 | 0.15 |
CHRM3-AS1 |
CHRM3 antisense RNA 1 |
83317 |
0.09 |
| chr8_59084985_59085323 | 0.15 |
FAM110B |
family with sequence similarity 110, member B |
178041 |
0.03 |
| chr1_229721184_229721335 | 0.15 |
ABCB10 |
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
26817 |
0.12 |
| chr12_53428271_53428519 | 0.15 |
RP11-983P16.4 |
|
11392 |
0.11 |
| chr2_29175143_29175407 | 0.15 |
FAM179A |
family with sequence similarity 179, member A |
4202 |
0.14 |
| chr14_77872559_77872757 | 0.15 |
FKSG61 |
|
10083 |
0.11 |
| chr13_53276312_53276509 | 0.15 |
LECT1 |
leukocyte cell derived chemotaxin 1 |
37077 |
0.16 |
| chr11_10646945_10647096 | 0.15 |
MRVI1 |
murine retrovirus integration site 1 homolog |
26712 |
0.15 |
| chr21_46955498_46955649 | 0.15 |
SLC19A1 |
solute carrier family 19 (folate transporter), member 1 |
1044 |
0.57 |
| chr6_18427248_18427399 | 0.15 |
ENSG00000238458 |
. |
25040 |
0.2 |
| chr3_30990444_30990685 | 0.15 |
GADL1 |
glutamate decarboxylase-like 1 |
54307 |
0.17 |
| chr1_17636167_17636318 | 0.15 |
PADI4 |
peptidyl arginine deiminase, type IV |
1550 |
0.36 |
| chr5_146165960_146166111 | 0.15 |
ENSG00000265875 |
. |
51637 |
0.15 |
| chr3_182929018_182929169 | 0.15 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
41939 |
0.13 |
| chr3_46134937_46135139 | 0.15 |
XCR1 |
chemokine (C motif) receptor 1 |
65804 |
0.1 |
| chr10_50568000_50568191 | 0.15 |
DRGX |
dorsal root ganglia homeobox |
31812 |
0.15 |
| chr20_23095516_23095667 | 0.15 |
CD93 |
CD93 molecule |
28614 |
0.15 |
| chr8_95983279_95983772 | 0.15 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
9636 |
0.15 |
| chr8_48418684_48418835 | 0.15 |
RP11-697N18.4 |
|
21200 |
0.24 |
| chr11_59571551_59571702 | 0.15 |
MRPL16 |
mitochondrial ribosomal protein L16 |
6147 |
0.16 |
| chr3_108988791_108989046 | 0.15 |
ENSG00000252889 |
. |
44942 |
0.15 |
| chr4_84115690_84115841 | 0.15 |
PLAC8 |
placenta-specific 8 |
57537 |
0.13 |
| chr7_117479656_117479807 | 0.15 |
CTTNBP2 |
cortactin binding protein 2 |
31961 |
0.24 |
| chr12_7596853_7597078 | 0.15 |
CD163L1 |
CD163 molecule-like 1 |
184 |
0.96 |
| chr1_199829495_199829849 | 0.15 |
ENSG00000200139 |
. |
27615 |
0.24 |
| chr5_157295124_157295340 | 0.15 |
CLINT1 |
clathrin interactor 1 |
9049 |
0.23 |
| chr3_184304717_184305002 | 0.15 |
EPHB3 |
EPH receptor B3 |
25287 |
0.17 |
| chr17_42761038_42761322 | 0.15 |
CCDC43 |
coiled-coil domain containing 43 |
5913 |
0.13 |
| chr5_73729078_73729239 | 0.15 |
ENSG00000244326 |
. |
115347 |
0.06 |
| chr2_176326738_176326889 | 0.15 |
ENSG00000221347 |
. |
131712 |
0.05 |
| chr18_11918822_11919203 | 0.14 |
MPPE1 |
metallophosphoesterase 1 |
9790 |
0.14 |
| chr16_81531821_81532117 | 0.14 |
CMIP |
c-Maf inducing protein |
3015 |
0.36 |
| chr22_50347685_50347933 | 0.14 |
PIM3 |
pim-3 oncogene |
6352 |
0.19 |
| chr1_68027658_68027809 | 0.14 |
ENSG00000207504 |
. |
20923 |
0.22 |
| chr7_50258904_50259055 | 0.14 |
AC020743.2 |
|
76560 |
0.1 |
| chr6_3139599_3139904 | 0.14 |
BPHL |
biphenyl hydrolase-like (serine hydrolase) |
908 |
0.5 |
| chr13_77390552_77390953 | 0.14 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
69773 |
0.11 |
| chr2_223907840_223908197 | 0.14 |
KCNE4 |
potassium voltage-gated channel, Isk-related family, member 4 |
8514 |
0.28 |
| chr2_54911007_54911158 | 0.14 |
AC093110.3 |
|
21153 |
0.19 |
| chr4_37394628_37394820 | 0.14 |
C4orf19 |
chromosome 4 open reading frame 19 |
60839 |
0.15 |
| chr6_6891304_6891524 | 0.14 |
ENSG00000240936 |
. |
47716 |
0.18 |
| chr6_13302166_13302317 | 0.14 |
RP1-257A7.4 |
|
6423 |
0.2 |
| chr7_6474034_6474265 | 0.14 |
DAGLB |
diacylglycerol lipase, beta |
13414 |
0.16 |
| chr5_88715552_88715703 | 0.14 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
1003 |
0.72 |
| chr19_52141106_52141257 | 0.14 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
7593 |
0.11 |
| chr7_97767382_97767616 | 0.14 |
LMTK2 |
lemur tyrosine kinase 2 |
31302 |
0.17 |
| chr1_41975943_41976221 | 0.14 |
RP11-486B10.3 |
|
25032 |
0.17 |
| chr1_95186096_95186247 | 0.14 |
ENSG00000263526 |
. |
25285 |
0.23 |
| chr2_169957115_169957325 | 0.14 |
AC007556.3 |
|
33 |
0.98 |
| chr9_93570502_93570735 | 0.14 |
SYK |
spleen tyrosine kinase |
6409 |
0.34 |
| chr3_14442673_14442824 | 0.14 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
1328 |
0.49 |
| chr14_93551916_93552209 | 0.14 |
ITPK1-AS1 |
ITPK1 antisense RNA 1 |
18265 |
0.18 |
| chr6_130012897_130013130 | 0.14 |
ARHGAP18 |
Rho GTPase activating protein 18 |
18357 |
0.25 |
| chr20_23128727_23129149 | 0.14 |
ENSG00000201527 |
. |
12670 |
0.21 |
| chr15_90562748_90563308 | 0.14 |
ENSG00000265871 |
. |
13041 |
0.14 |
| chr1_32123678_32123829 | 0.14 |
RP11-73M7.6 |
|
894 |
0.46 |
| chr3_184293955_184294106 | 0.14 |
EPHB3 |
EPH receptor B3 |
14458 |
0.18 |
| chr10_126758424_126758575 | 0.14 |
ENSG00000264572 |
. |
37060 |
0.17 |
| chr4_106035810_106035961 | 0.14 |
ENSG00000252136 |
. |
8741 |
0.23 |
| chr11_15923291_15923597 | 0.14 |
CTD-3096P4.1 |
|
121292 |
0.06 |
| chr15_43522743_43522938 | 0.14 |
EPB42 |
erythrocyte membrane protein band 4.2 |
9359 |
0.13 |
| chr8_56887528_56887788 | 0.14 |
ENSG00000240905 |
. |
5438 |
0.18 |
| chr15_50698058_50698434 | 0.14 |
USP8 |
ubiquitin specific peptidase 8 |
18331 |
0.14 |
| chr2_38493865_38494016 | 0.14 |
ATL2 |
atlastin GTPase 2 |
109426 |
0.06 |
| chr3_196280624_196280799 | 0.13 |
WDR53 |
WD repeat domain 53 |
13470 |
0.13 |
| chr8_96220752_96221005 | 0.13 |
C8orf37 |
chromosome 8 open reading frame 37 |
60551 |
0.12 |
| chr1_224097972_224098123 | 0.13 |
TP53BP2 |
tumor protein p53 binding protein, 2 |
64373 |
0.11 |
| chr7_69368156_69368307 | 0.13 |
AUTS2 |
autism susceptibility candidate 2 |
303634 |
0.01 |
| chr6_36524306_36524457 | 0.13 |
STK38 |
serine/threonine kinase 38 |
9134 |
0.15 |
| chr20_2265960_2266140 | 0.13 |
TGM3 |
transglutaminase 3 |
10597 |
0.24 |
| chr5_159743183_159743334 | 0.13 |
CCNJL |
cyclin J-like |
3651 |
0.22 |
| chr5_3994179_3994446 | 0.13 |
CTD-2012M11.3 |
|
394010 |
0.01 |
| chr3_15973999_15974271 | 0.13 |
ENSG00000207815 |
. |
58857 |
0.13 |
| chr3_45456350_45456693 | 0.13 |
LARS2 |
leucyl-tRNA synthetase 2, mitochondrial |
26410 |
0.21 |
| chr4_22609177_22609328 | 0.13 |
GPR125 |
G protein-coupled receptor 125 |
91575 |
0.1 |
| chr12_29400868_29401019 | 0.13 |
FAR2 |
fatty acyl CoA reductase 2 |
24265 |
0.22 |
| chr19_43057216_43057430 | 0.13 |
CEACAM1 |
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
8063 |
0.19 |
| chr9_78221708_78221880 | 0.13 |
ENSG00000238598 |
. |
70029 |
0.13 |
| chr9_94773267_94773418 | 0.13 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
60898 |
0.13 |
| chr20_2266440_2266591 | 0.13 |
TGM3 |
transglutaminase 3 |
10132 |
0.24 |
| chr19_43849836_43850167 | 0.13 |
CD177 |
CD177 molecule |
7824 |
0.17 |
| chr3_67047069_67047341 | 0.13 |
KBTBD8 |
kelch repeat and BTB (POZ) domain containing 8 |
1522 |
0.59 |
| chr4_39152584_39152984 | 0.13 |
RP11-360F5.1 |
|
24346 |
0.17 |
| chr12_54688907_54689323 | 0.13 |
NFE2 |
nuclear factor, erythroid 2 |
427 |
0.65 |
| chr1_10748665_10749049 | 0.13 |
CASZ1 |
castor zinc finger 1 |
43724 |
0.14 |
| chr15_89710472_89711584 | 0.13 |
RLBP1 |
retinaldehyde binding protein 1 |
44046 |
0.12 |
| chr2_25706012_25706163 | 0.13 |
AC104699.1 |
|
62101 |
0.13 |
| chrX_39667886_39668149 | 0.13 |
ENSG00000264618 |
. |
21940 |
0.22 |
| chr14_24041269_24041503 | 0.13 |
JPH4 |
junctophilin 4 |
824 |
0.44 |
| chr12_89555329_89555814 | 0.13 |
ENSG00000238302 |
. |
120491 |
0.06 |
| chr7_39740325_39740528 | 0.13 |
AC004837.5 |
|
2469 |
0.34 |
| chr11_126041739_126041935 | 0.13 |
RP11-50B3.4 |
|
37085 |
0.1 |
| chr3_16006227_16006629 | 0.13 |
ENSG00000207815 |
. |
91150 |
0.08 |
| chr17_56324893_56325185 | 0.13 |
LPO |
lactoperoxidase |
8963 |
0.13 |
| chr20_58688912_58689063 | 0.13 |
C20orf197 |
chromosome 20 open reading frame 197 |
58007 |
0.14 |
| chr2_121946677_121947036 | 0.13 |
TFCP2L1 |
transcription factor CP2-like 1 |
95927 |
0.08 |
| chr2_192301147_192301752 | 0.13 |
MYO1B |
myosin IB |
25658 |
0.23 |
| chr17_47590468_47590727 | 0.13 |
NGFR |
nerve growth factor receptor |
15844 |
0.14 |
| chr2_111790938_111791089 | 0.13 |
ACOXL |
acyl-CoA oxidase-like |
46238 |
0.17 |
| chr11_122263966_122264117 | 0.13 |
ENSG00000252776 |
. |
55877 |
0.13 |
| chr3_128312961_128313112 | 0.13 |
C3orf27 |
chromosome 3 open reading frame 27 |
18107 |
0.19 |
| chr9_73196230_73196387 | 0.13 |
ENSG00000272232 |
. |
1931 |
0.46 |
| chr4_36313792_36313943 | 0.13 |
DTHD1 |
death domain containing 1 |
28223 |
0.2 |
| chr1_247805569_247805720 | 0.13 |
RP11-978I15.10 |
|
1488 |
0.25 |
| chr9_126319054_126319231 | 0.13 |
ENSG00000222722 |
. |
28594 |
0.2 |
| chr8_86983503_86983666 | 0.13 |
ATP6V0D2 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
15968 |
0.26 |
| chr1_29111482_29111776 | 0.13 |
OPRD1 |
opioid receptor, delta 1 |
27025 |
0.15 |
| chr10_101939847_101940043 | 0.13 |
ERLIN1 |
ER lipid raft associated 1 |
5841 |
0.15 |
| chr7_41909046_41909309 | 0.13 |
AC005027.3 |
|
164244 |
0.04 |
| chr1_26601413_26601689 | 0.13 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
4116 |
0.14 |
| chr9_124222243_124222394 | 0.13 |
ENSG00000240299 |
. |
34479 |
0.15 |
| chr9_137248256_137248507 | 0.13 |
ENSG00000263897 |
. |
22876 |
0.21 |
| chr2_178213706_178213857 | 0.13 |
ENSG00000238295 |
. |
3092 |
0.19 |
| chr4_139875353_139875683 | 0.13 |
RP11-371F15.3 |
|
16383 |
0.2 |
| chr2_89053827_89053978 | 0.13 |
ENSG00000221686 |
. |
12933 |
0.13 |
| chr2_240072595_240072746 | 0.13 |
HDAC4 |
histone deacetylase 4 |
4601 |
0.15 |
| chr9_92771423_92771933 | 0.13 |
ENSG00000263967 |
. |
14139 |
0.32 |
| chr1_229294714_229294972 | 0.13 |
RP5-1061H20.5 |
|
68466 |
0.1 |
| chr17_8693564_8693757 | 0.13 |
MFSD6L |
major facilitator superfamily domain containing 6-like |
9007 |
0.19 |
| chr6_35002781_35002932 | 0.13 |
TCP11 |
t-complex 11, testis-specific |
85966 |
0.08 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.3 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 | 0.2 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.1 | 0.2 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.3 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.1 | 0.1 | GO:0052555 | positive regulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052555) positive regulation by symbiont of host immune response(GO:0052556) |
| 0.1 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.2 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.1 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.2 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.0 | GO:0032621 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.2 | GO:0009415 | response to water(GO:0009415) |
| 0.0 | 0.2 | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.2 | GO:0046881 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.2 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.3 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.4 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.0 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.1 | GO:0032352 | positive regulation of hormone metabolic process(GO:0032352) positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.2 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.2 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.2 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.0 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.0 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.0 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.0 | 0.0 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.0 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.2 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.0 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.2 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.0 | GO:0051883 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.0 | GO:0051957 | positive regulation of amino acid transport(GO:0051957) |
| 0.0 | 0.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.0 | GO:0050686 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0032329 | serine transport(GO:0032329) |
| 0.0 | 0.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0033866 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.1 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.0 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.0 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0009595 | detection of biotic stimulus(GO:0009595) |
| 0.0 | 0.1 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.0 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.0 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.0 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.0 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.0 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0031082 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.6 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.0 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.0 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.0 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0015645 | fatty acid ligase activity(GO:0015645) |
| 0.0 | 0.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.0 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.0 | REACTOME MITOTIC G1 G1 S PHASES | Genes involved in Mitotic G1-G1/S phases |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |