Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
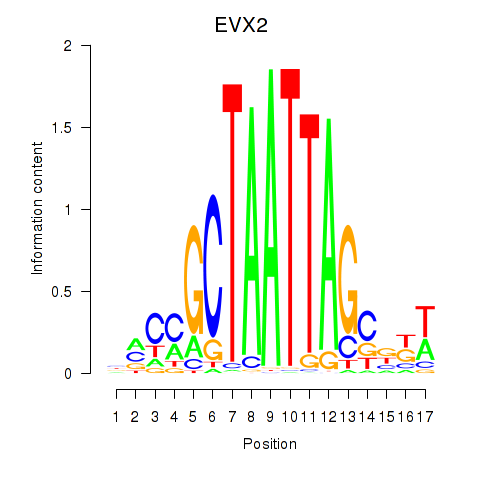
Results for EVX2
Z-value: 0.99
Transcription factors associated with EVX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EVX2
|
ENSG00000174279.4 | even-skipped homeobox 2 |
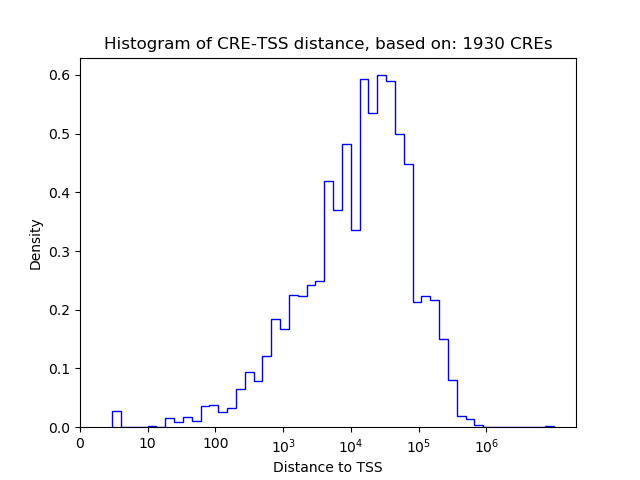
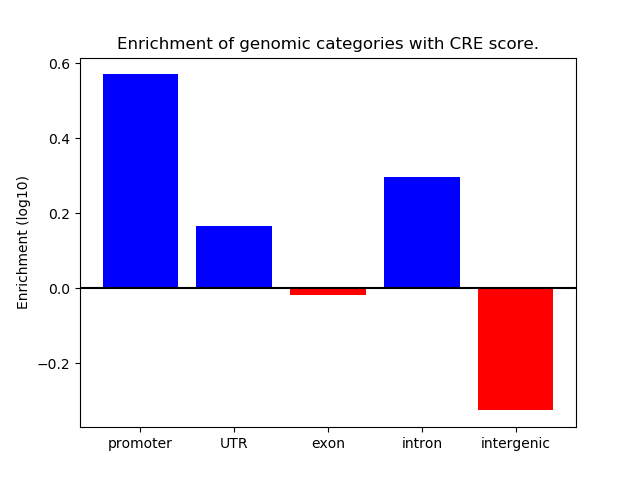
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_176932380_176932585 | EVX2 | 16159 | 0.098495 | 0.78 | 1.4e-02 | Click! |
| chr2_176945583_176945734 | EVX2 | 2983 | 0.131117 | 0.76 | 1.7e-02 | Click! |
| chr2_176947944_176948095 | EVX2 | 622 | 0.538477 | 0.74 | 2.2e-02 | Click! |
| chr2_176950582_176950733 | EVX2 | 2016 | 0.164980 | 0.73 | 2.6e-02 | Click! |
| chr2_176946726_176946982 | EVX2 | 1787 | 0.191724 | 0.72 | 3.0e-02 | Click! |
Activity of the EVX2 motif across conditions
Conditions sorted by the z-value of the EVX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
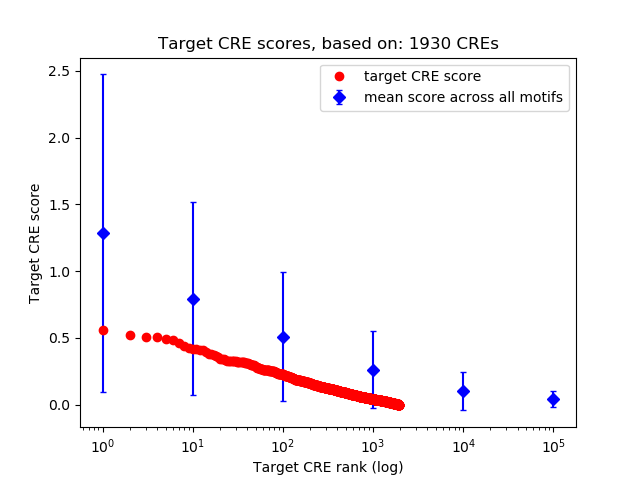
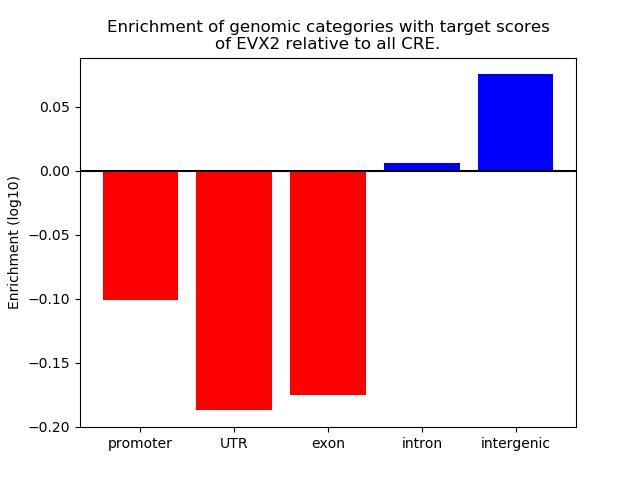
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_174862628_174862955 | 0.56 |
SP3 |
Sp3 transcription factor |
32361 |
0.24 |
| chr10_79021361_79021706 | 0.52 |
RP11-328K22.1 |
|
52166 |
0.15 |
| chr7_41910301_41910559 | 0.51 |
AC005027.3 |
|
165497 |
0.04 |
| chr1_173159483_173159962 | 0.51 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
14975 |
0.26 |
| chr17_26498050_26498459 | 0.49 |
PYY2 |
peptide YY, 2 (pseudogene) |
56078 |
0.09 |
| chr11_10609627_10609795 | 0.49 |
MRVI1-AS1 |
MRVI1 antisense RNA 1 |
5033 |
0.19 |
| chr18_54041028_54041261 | 0.46 |
TXNL1 |
thioredoxin-like 1 |
240578 |
0.02 |
| chr2_86017810_86018001 | 0.44 |
AC105053.3 |
|
24348 |
0.13 |
| chr20_40221307_40221497 | 0.42 |
CHD6 |
chromodomain helicase DNA binding protein 6 |
22010 |
0.23 |
| chr3_12230073_12230226 | 0.42 |
TIMP4 |
TIMP metallopeptidase inhibitor 4 |
29298 |
0.22 |
| chr13_33222085_33222352 | 0.41 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
61654 |
0.13 |
| chr3_185538752_185538943 | 0.41 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
2 |
0.98 |
| chr15_67006142_67006408 | 0.41 |
SMAD6 |
SMAD family member 6 |
2240 |
0.39 |
| chrX_38728176_38728358 | 0.40 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
65131 |
0.13 |
| chr2_64881941_64882849 | 0.38 |
SERTAD2 |
SERTA domain containing 2 |
1348 |
0.52 |
| chr2_163157595_163157835 | 0.38 |
IFIH1 |
interferon induced with helicase C domain 1 |
17479 |
0.19 |
| chr3_5469487_5469638 | 0.37 |
ENSG00000241227 |
. |
174654 |
0.03 |
| chr16_85295971_85296292 | 0.36 |
ENSG00000266307 |
. |
43800 |
0.16 |
| chr5_138483370_138483521 | 0.36 |
SIL1 |
SIL1 nucleotide exchange factor |
7711 |
0.22 |
| chr5_111109748_111109899 | 0.34 |
NREP |
neuronal regeneration related protein |
15908 |
0.22 |
| chr10_95236478_95236629 | 0.34 |
MYOF |
myoferlin |
5398 |
0.21 |
| chr10_102813488_102813726 | 0.34 |
KAZALD1 |
Kazal-type serine peptidase inhibitor domain 1 |
7991 |
0.12 |
| chr9_138952025_138952443 | 0.34 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
9808 |
0.22 |
| chr8_121665009_121665299 | 0.33 |
RP11-713M15.1 |
|
108339 |
0.07 |
| chr7_20071186_20071471 | 0.33 |
AC005062.2 |
|
28783 |
0.23 |
| chr1_28797868_28798297 | 0.33 |
ENSG00000221216 |
. |
9792 |
0.12 |
| chr1_196578004_196578155 | 0.33 |
KCNT2 |
potassium channel, subfamily T, member 2 |
276 |
0.93 |
| chr1_100976278_100976464 | 0.33 |
RP5-837M10.4 |
|
24818 |
0.19 |
| chr11_122129149_122129397 | 0.33 |
RP11-716H6.2 |
|
36625 |
0.18 |
| chr2_38046333_38046643 | 0.32 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
80877 |
0.1 |
| chr5_112314540_112314973 | 0.32 |
DCP2 |
decapping mRNA 2 |
2277 |
0.32 |
| chr18_72443447_72443598 | 0.32 |
ZNF407 |
zinc finger protein 407 |
100546 |
0.08 |
| chr8_142158040_142158331 | 0.32 |
DENND3 |
DENN/MADD domain containing 3 |
6883 |
0.21 |
| chr20_50419228_50419379 | 0.32 |
SALL4 |
spalt-like transcription factor 4 |
244 |
0.94 |
| chr3_71862606_71862965 | 0.32 |
ENSG00000239250 |
. |
13543 |
0.2 |
| chr4_74571517_74571668 | 0.32 |
IL8 |
interleukin 8 |
34631 |
0.18 |
| chr4_124476344_124476624 | 0.32 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
155361 |
0.04 |
| chrX_10135860_10136222 | 0.31 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
9469 |
0.24 |
| chr6_15349427_15349643 | 0.31 |
ENSG00000201519 |
. |
24832 |
0.19 |
| chr12_111888065_111888216 | 0.31 |
ATXN2 |
ataxin 2 |
5776 |
0.19 |
| chr12_1613825_1614060 | 0.31 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
25115 |
0.21 |
| chr3_148896549_148896700 | 0.31 |
CP |
ceruloplasmin (ferroxidase) |
784 |
0.67 |
| chr7_135656817_135657003 | 0.30 |
MTPN |
myotrophin |
5154 |
0.19 |
| chr11_61737024_61737182 | 0.30 |
AP003733.1 |
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
1650 |
0.25 |
| chr6_15897774_15897925 | 0.30 |
MYLIP |
myosin regulatory light chain interacting protein |
231507 |
0.02 |
| chr4_130455151_130455302 | 0.30 |
ENSG00000222861 |
. |
326429 |
0.01 |
| chr2_71503166_71503343 | 0.30 |
PAIP2B |
poly(A) binding protein interacting protein 2B |
49041 |
0.12 |
| chr6_37669094_37669483 | 0.30 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
2206 |
0.4 |
| chr5_43208877_43209028 | 0.29 |
NIM1K |
NIM1 serine/threonine protein kinase |
15998 |
0.21 |
| chr3_159902915_159903066 | 0.28 |
IL12A-AS1 |
IL12A antisense RNA 1 |
21889 |
0.18 |
| chr18_74872232_74872572 | 0.28 |
MBP |
myelin basic protein |
27602 |
0.24 |
| chr7_148320637_148320788 | 0.28 |
C7orf33 |
chromosome 7 open reading frame 33 |
33055 |
0.18 |
| chr13_33836579_33836730 | 0.28 |
ENSG00000236581 |
. |
8862 |
0.23 |
| chr11_12767798_12767949 | 0.27 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
1285 |
0.56 |
| chr11_94315586_94315737 | 0.27 |
PIWIL4 |
piwi-like RNA-mediated gene silencing 4 |
14804 |
0.19 |
| chr22_38795449_38796163 | 0.27 |
CSNK1E |
casein kinase 1, epsilon |
1279 |
0.34 |
| chr2_166451082_166451288 | 0.27 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
20566 |
0.27 |
| chr2_235485735_235485886 | 0.27 |
ARL4C |
ADP-ribosylation factor-like 4C |
80113 |
0.12 |
| chr13_77319833_77320172 | 0.26 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
140523 |
0.05 |
| chr16_53133154_53133539 | 0.26 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
274 |
0.94 |
| chr2_208116387_208116719 | 0.26 |
AC007879.5 |
|
2423 |
0.32 |
| chr12_62525076_62525421 | 0.26 |
FAM19A2 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
4569 |
0.22 |
| chr2_131100080_131100231 | 0.26 |
CCDC115 |
coiled-coil domain containing 115 |
233 |
0.56 |
| chr2_216768664_216768815 | 0.26 |
ENSG00000212055 |
. |
25097 |
0.24 |
| chr16_85296592_85296743 | 0.26 |
ENSG00000266307 |
. |
43264 |
0.16 |
| chr11_69264697_69264848 | 0.26 |
CCND1 |
cyclin D1 |
191083 |
0.03 |
| chr8_105598551_105598718 | 0.26 |
LRP12 |
low density lipoprotein receptor-related protein 12 |
2600 |
0.3 |
| chr13_60026787_60027144 | 0.26 |
ENSG00000239003 |
. |
27239 |
0.28 |
| chr5_17413551_17413702 | 0.26 |
ENSG00000201715 |
. |
67901 |
0.13 |
| chr10_78983856_78984007 | 0.26 |
RP11-180I22.2 |
|
75881 |
0.1 |
| chr7_75952019_75952170 | 0.25 |
HSPB1 |
heat shock 27kDa protein 1 |
19200 |
0.14 |
| chr6_5828101_5828316 | 0.25 |
ENSG00000239472 |
. |
90869 |
0.09 |
| chr2_112368076_112368227 | 0.25 |
ENSG00000266063 |
. |
160560 |
0.04 |
| chr13_88323755_88324230 | 0.25 |
SLITRK5 |
SLIT and NTRK-like family, member 5 |
878 |
0.73 |
| chr19_7268375_7268570 | 0.25 |
INSR |
insulin receptor |
25470 |
0.17 |
| chr1_243896013_243896164 | 0.25 |
RP11-370K11.1 |
|
8035 |
0.27 |
| chr9_95298761_95298917 | 0.25 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
98 |
0.97 |
| chr21_42579078_42579229 | 0.25 |
PLAC4 |
placenta-specific 4 |
20438 |
0.15 |
| chr10_129704503_129704722 | 0.25 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
713 |
0.71 |
| chr14_99502782_99502941 | 0.25 |
AL162151.4 |
|
121892 |
0.06 |
| chr8_130906812_130906977 | 0.25 |
ENSG00000200075 |
. |
25932 |
0.18 |
| chr1_185066313_185066464 | 0.24 |
TRMT1L |
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
42304 |
0.13 |
| chr9_110522175_110522455 | 0.24 |
AL162389.1 |
Uncharacterized protein |
18104 |
0.2 |
| chr20_52352229_52352983 | 0.24 |
ENSG00000238468 |
. |
67309 |
0.12 |
| chr1_206225703_206225865 | 0.24 |
AVPR1B |
arginine vasopressin receptor 1B |
1808 |
0.28 |
| chr4_128619906_128620188 | 0.24 |
INTU |
inturned planar cell polarity protein |
11078 |
0.23 |
| chr7_23030019_23030170 | 0.24 |
FAM126A |
family with sequence similarity 126, member A |
23599 |
0.23 |
| chr6_12940320_12940471 | 0.23 |
PHACTR1 |
phosphatase and actin regulator 1 |
17819 |
0.29 |
| chr1_116274836_116274987 | 0.23 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
36424 |
0.18 |
| chr1_225610187_225610435 | 0.23 |
LBR |
lamin B receptor |
5288 |
0.2 |
| chr8_38238060_38238354 | 0.23 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
132 |
0.94 |
| chr17_33873153_33873675 | 0.23 |
RP11-1094M14.12 |
|
6810 |
0.11 |
| chr8_22949674_22949892 | 0.23 |
TNFRSF10C |
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain |
7915 |
0.13 |
| chr2_55390382_55390533 | 0.23 |
RTN4 |
reticulon 4 |
50700 |
0.1 |
| chr10_131682317_131682501 | 0.23 |
ENSG00000266676 |
. |
40771 |
0.18 |
| chr18_3015862_3016088 | 0.23 |
LPIN2 |
lipin 2 |
2662 |
0.24 |
| chr3_151066517_151066717 | 0.23 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
19281 |
0.16 |
| chr2_168048837_168048988 | 0.23 |
XIRP2 |
xin actin-binding repeat containing 2 |
5119 |
0.31 |
| chr6_35014782_35015029 | 0.23 |
TCP11 |
t-complex 11, testis-specific |
73917 |
0.09 |
| chr13_102302331_102302482 | 0.23 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
51911 |
0.17 |
| chr10_15921921_15922072 | 0.23 |
ENSG00000239130 |
. |
6361 |
0.25 |
| chr17_62384463_62384614 | 0.22 |
TEX2 |
testis expressed 2 |
43877 |
0.13 |
| chr1_238770348_238770644 | 0.22 |
ENSG00000252371 |
. |
281408 |
0.01 |
| chr8_99838628_99838779 | 0.22 |
STK3 |
serine/threonine kinase 3 |
794 |
0.74 |
| chr6_30923154_30923376 | 0.22 |
HCG21 |
HLA complex group 21 (non-protein coding) |
626 |
0.52 |
| chr5_4199619_4199770 | 0.22 |
CTD-2012M11.3 |
|
599392 |
0.0 |
| chr6_161525948_161526099 | 0.22 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
44298 |
0.18 |
| chr6_134238115_134238266 | 0.22 |
RP3-323P13.2 |
|
25116 |
0.18 |
| chr2_161236579_161237045 | 0.22 |
ENSG00000252465 |
. |
16667 |
0.2 |
| chr5_73839209_73839360 | 0.22 |
HEXB |
hexosaminidase B (beta polypeptide) |
96564 |
0.07 |
| chr8_122257591_122257742 | 0.21 |
ENSG00000221644 |
. |
58534 |
0.16 |
| chr2_44576037_44576404 | 0.21 |
PREPL |
prolyl endopeptidase-like |
10668 |
0.2 |
| chr1_8271781_8272061 | 0.21 |
ENSG00000200975 |
. |
5264 |
0.25 |
| chr22_24908185_24908422 | 0.21 |
AP000355.2 |
|
4173 |
0.16 |
| chr4_95454799_95455193 | 0.21 |
PDLIM5 |
PDZ and LIM domain 5 |
10120 |
0.32 |
| chr5_135394625_135394981 | 0.21 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
72 |
0.98 |
| chr2_159955686_159955837 | 0.21 |
ENSG00000202029 |
. |
72107 |
0.11 |
| chr1_9301737_9301925 | 0.21 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
1928 |
0.34 |
| chr7_16851610_16851761 | 0.21 |
AGR2 |
anterior gradient 2 |
6981 |
0.2 |
| chr3_111717564_111717859 | 0.21 |
TAGLN3 |
transgelin 3 |
69 |
0.97 |
| chr5_136899227_136899378 | 0.20 |
ENSG00000221612 |
. |
30159 |
0.2 |
| chr4_139943173_139943324 | 0.20 |
ENSG00000264953 |
. |
4515 |
0.17 |
| chr7_29247042_29247209 | 0.20 |
AC004593.3 |
|
1461 |
0.41 |
| chr14_101178681_101178832 | 0.20 |
DLK1 |
delta-like 1 homolog (Drosophila) |
13286 |
0.12 |
| chr19_954292_954443 | 0.20 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
21858 |
0.07 |
| chr14_105749762_105749913 | 0.20 |
BRF1 |
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
16968 |
0.15 |
| chr1_231493802_231494072 | 0.20 |
SPRTN |
SprT-like N-terminal domain |
18764 |
0.17 |
| chr8_19213262_19213413 | 0.20 |
SH2D4A |
SH2 domain containing 4A |
36236 |
0.23 |
| chr14_60671402_60671553 | 0.20 |
DHRS7 |
dehydrogenase/reductase (SDR family) member 7 |
34903 |
0.15 |
| chr7_138660539_138660690 | 0.20 |
KIAA1549 |
KIAA1549 |
5450 |
0.26 |
| chr4_120580539_120580789 | 0.20 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
30518 |
0.22 |
| chr6_56591218_56591369 | 0.19 |
DST |
dystonin |
59384 |
0.15 |
| chr2_102914112_102914534 | 0.19 |
IL1RL1 |
interleukin 1 receptor-like 1 |
13639 |
0.19 |
| chr3_137930306_137930457 | 0.19 |
ARMC8 |
armadillo repeat containing 8 |
12097 |
0.21 |
| chr22_29641192_29641343 | 0.19 |
EMID1 |
EMI domain containing 1 |
14260 |
0.11 |
| chr5_150861749_150861900 | 0.19 |
ENSG00000200227 |
. |
4686 |
0.19 |
| chr8_70583516_70583667 | 0.19 |
ENSG00000222889 |
. |
28868 |
0.19 |
| chr12_65369994_65370192 | 0.19 |
ENSG00000221564 |
. |
37290 |
0.18 |
| chr6_26485415_26485566 | 0.19 |
BTN1A1 |
butyrophilin, subfamily 1, member A1 |
15959 |
0.11 |
| chr9_128517768_128517919 | 0.19 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
7365 |
0.28 |
| chr8_33325166_33325317 | 0.19 |
FUT10 |
fucosyltransferase 10 (alpha (1,3) fucosyltransferase) |
5396 |
0.18 |
| chr5_34519672_34519823 | 0.19 |
RAI14 |
retinoic acid induced 14 |
136595 |
0.05 |
| chr7_81620419_81620570 | 0.19 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
14612 |
0.22 |
| chr14_54913534_54913685 | 0.19 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
5460 |
0.23 |
| chr6_157758656_157758807 | 0.19 |
TMEM242 |
transmembrane protein 242 |
14098 |
0.22 |
| chr3_155926514_155926665 | 0.19 |
KCNAB1-AS2 |
KCNAB1 antisense RNA 2 |
19083 |
0.25 |
| chr17_64467443_64467600 | 0.19 |
RP11-4F22.2 |
|
54549 |
0.14 |
| chr17_59327352_59327704 | 0.18 |
RP11-136H19.1 |
|
114203 |
0.05 |
| chr14_77873001_77873391 | 0.18 |
FKSG61 |
|
9545 |
0.11 |
| chr15_56439129_56439280 | 0.18 |
RFX7 |
regulatory factor X, 7 |
42880 |
0.17 |
| chr5_139090606_139090882 | 0.18 |
ENSG00000200756 |
. |
6931 |
0.21 |
| chr4_173934863_173935106 | 0.18 |
ENSG00000241652 |
. |
127810 |
0.05 |
| chr2_18733477_18733628 | 0.18 |
RDH14 |
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
8394 |
0.2 |
| chr14_103046146_103046463 | 0.18 |
ENSG00000239224 |
. |
3210 |
0.22 |
| chr9_110473577_110473728 | 0.18 |
ENSG00000202308 |
. |
48125 |
0.15 |
| chr4_186629734_186630027 | 0.18 |
ENSG00000207497 |
. |
1241 |
0.48 |
| chr4_26220381_26220779 | 0.18 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
53649 |
0.18 |
| chr6_146866872_146867091 | 0.18 |
RAB32 |
RAB32, member RAS oncogene family |
2152 |
0.41 |
| chr5_74615457_74615613 | 0.18 |
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
16619 |
0.19 |
| chr3_105699405_105699556 | 0.18 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
111084 |
0.08 |
| chr22_25799352_25799559 | 0.18 |
LRP5L |
low density lipoprotein receptor-related protein 5-like |
1889 |
0.35 |
| chr3_24630898_24631049 | 0.18 |
ENSG00000265028 |
. |
68047 |
0.12 |
| chr11_75561572_75561825 | 0.18 |
UVRAG |
UV radiation resistance associated |
11709 |
0.12 |
| chr10_104557046_104557197 | 0.18 |
ENSG00000252994 |
. |
6565 |
0.14 |
| chr5_173293788_173294026 | 0.18 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
21376 |
0.25 |
| chr13_27597624_27597775 | 0.18 |
USP12-AS1 |
USP12 antisense RNA 1 |
139293 |
0.04 |
| chr12_65684698_65684849 | 0.18 |
RP11-305O6.3 |
|
9336 |
0.23 |
| chr1_116185862_116186059 | 0.18 |
VANGL1 |
VANGL planar cell polarity protein 1 |
710 |
0.71 |
| chr12_32609742_32609949 | 0.18 |
ENSG00000253063 |
. |
20560 |
0.2 |
| chr11_114129904_114130134 | 0.17 |
NNMT |
nicotinamide N-methyltransferase |
1466 |
0.5 |
| chr12_25653663_25653956 | 0.17 |
IFLTD1 |
intermediate filament tail domain containing 1 |
19086 |
0.25 |
| chr2_63926133_63926284 | 0.17 |
ENSG00000221085 |
. |
3539 |
0.34 |
| chr14_89633575_89633726 | 0.17 |
FOXN3 |
forkhead box N3 |
13438 |
0.28 |
| chr10_173434_173996 | 0.17 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
6690 |
0.24 |
| chr11_112193182_112193453 | 0.17 |
ENSG00000206772 |
. |
30068 |
0.14 |
| chr9_16828594_16828745 | 0.17 |
BNC2 |
basonuclin 2 |
3617 |
0.36 |
| chr6_114017168_114017357 | 0.17 |
ENSG00000221559 |
. |
8773 |
0.26 |
| chr11_117958729_117958938 | 0.17 |
TMPRSS4-AS1 |
TMPRSS4 antisense RNA 1 |
1325 |
0.38 |
| chr12_89033164_89033405 | 0.17 |
ENSG00000252850 |
. |
57868 |
0.14 |
| chr3_116583241_116583392 | 0.17 |
ENSG00000265433 |
. |
14102 |
0.25 |
| chr3_111623515_111623944 | 0.17 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
6728 |
0.26 |
| chr19_42315524_42315845 | 0.17 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
14599 |
0.11 |
| chr17_58383468_58383619 | 0.17 |
USP32 |
ubiquitin specific peptidase 32 |
49482 |
0.13 |
| chr20_52211917_52212263 | 0.17 |
ZNF217 |
zinc finger protein 217 |
1712 |
0.38 |
| chr1_33115546_33115708 | 0.17 |
ZBTB8OS |
zinc finger and BTB domain containing 8 opposite strand |
534 |
0.62 |
| chrX_131614461_131614831 | 0.17 |
MBNL3 |
muscleblind-like splicing regulator 3 |
8397 |
0.3 |
| chr21_30503482_30503633 | 0.17 |
MAP3K7CL |
MAP3K7 C-terminal like |
21 |
0.97 |
| chr11_88078061_88078240 | 0.17 |
CTSC |
cathepsin C |
7195 |
0.31 |
| chr6_20484087_20484358 | 0.17 |
ENSG00000201683 |
. |
16184 |
0.16 |
| chr1_178102702_178103151 | 0.17 |
RASAL2 |
RAS protein activator like 2 |
39650 |
0.22 |
| chr20_48955704_48955952 | 0.17 |
ENSG00000244376 |
. |
90192 |
0.08 |
| chr5_123149843_123149994 | 0.17 |
CSNK1G3 |
casein kinase 1, gamma 3 |
225799 |
0.02 |
| chr1_205206462_205206644 | 0.17 |
RP11-383G10.5 |
|
558 |
0.73 |
| chr6_11912233_11912384 | 0.16 |
RP11-456H18.1 |
|
79937 |
0.1 |
| chr17_47440332_47440604 | 0.16 |
ZNF652 |
zinc finger protein 652 |
633 |
0.61 |
| chr12_56027127_56027278 | 0.16 |
OR10P1 |
olfactory receptor, family 10, subfamily P, member 1 |
3442 |
0.14 |
| chr3_99764961_99765177 | 0.16 |
FILIP1L |
filamin A interacting protein 1-like |
68288 |
0.1 |
| chr2_144712590_144712741 | 0.16 |
AC016910.1 |
|
18025 |
0.25 |
| chr8_80226395_80226546 | 0.16 |
ENSG00000264969 |
. |
216872 |
0.02 |
| chr5_126306662_126306822 | 0.16 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
52822 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.0 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.0 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.0 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.0 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.0 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME SIGNALLING TO ERKS | Genes involved in Signalling to ERKs |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |