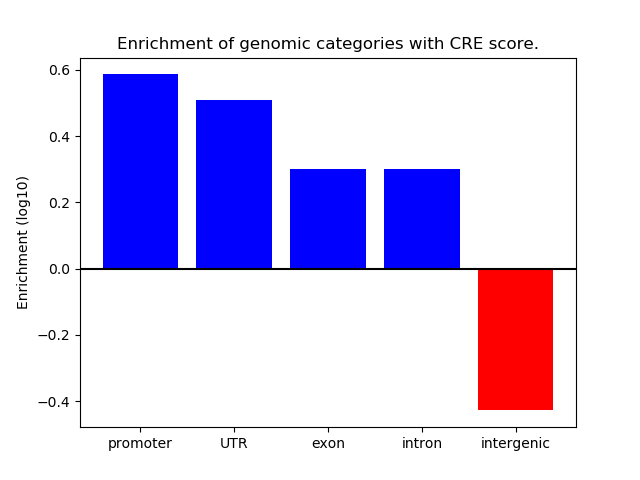
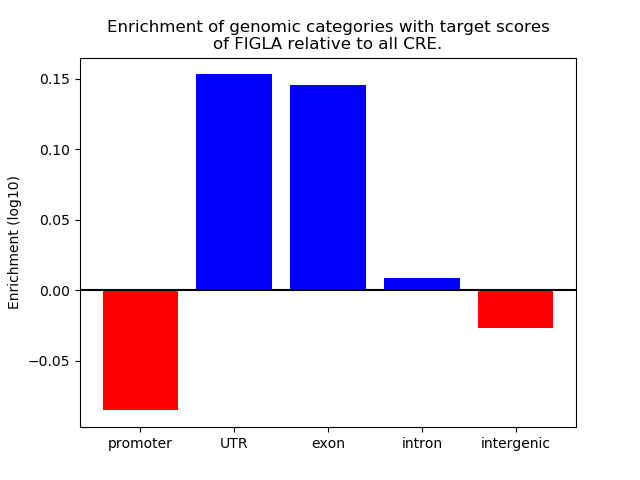
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
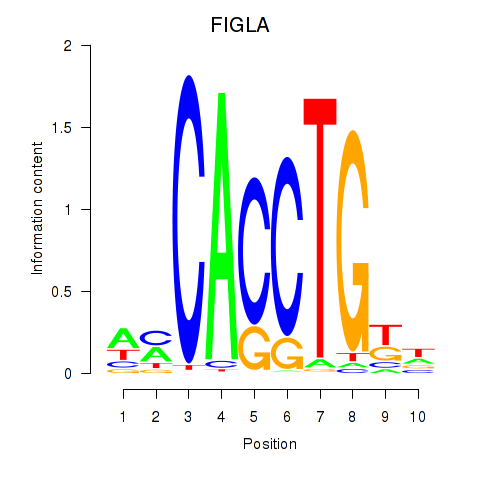
Results for FIGLA
Z-value: 0.75
Transcription factors associated with FIGLA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FIGLA
|
ENSG00000183733.6 | folliculogenesis specific bHLH transcription factor |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_71016692_71017138 | FIGLA | 860 | 0.548680 | -0.49 | 1.8e-01 | Click! |
| chr2_71020635_71020786 | FIGLA | 2935 | 0.209139 | 0.29 | 4.4e-01 | Click! |
| chr2_71017167_71017584 | FIGLA | 400 | 0.818461 | -0.19 | 6.2e-01 | Click! |
| chr2_71020886_71021037 | FIGLA | 3186 | 0.200287 | 0.02 | 9.7e-01 | Click! |
Activity of the FIGLA motif across conditions
Conditions sorted by the z-value of the FIGLA motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
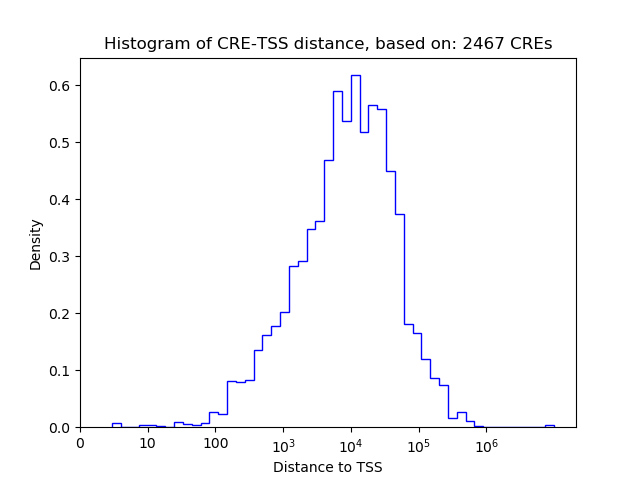
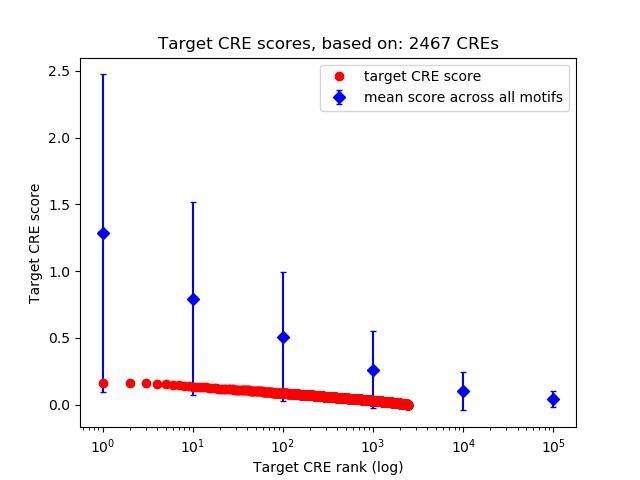
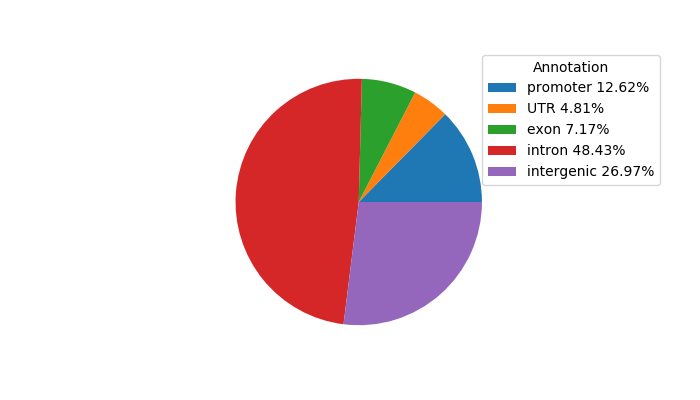
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_68107852_68108775 | 0.16 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
10934 |
0.1 |
| chr8_129254431_129254830 | 0.16 |
ENSG00000201782 |
. |
21880 |
0.26 |
| chr2_43402387_43402951 | 0.16 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
51079 |
0.14 |
| chr9_95724443_95724972 | 0.16 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
1536 |
0.44 |
| chr12_95045333_95045484 | 0.16 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
1070 |
0.64 |
| chr2_219182043_219182360 | 0.15 |
PNKD |
paroxysmal nonkinesigenic dyskinesia |
5695 |
0.11 |
| chr1_236040463_236040645 | 0.14 |
LYST |
lysosomal trafficking regulator |
6318 |
0.17 |
| chr18_74794512_74794972 | 0.14 |
MBP |
myelin basic protein |
22475 |
0.26 |
| chr14_106559690_106559937 | 0.14 |
IGHV2-10 |
immunoglobulin heavy variable 2-10 (pseudogene) |
162 |
0.82 |
| chr1_206737001_206737160 | 0.14 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
6587 |
0.17 |
| chr1_169673193_169673400 | 0.13 |
SELL |
selectin L |
7543 |
0.2 |
| chr11_119558435_119558586 | 0.13 |
ENSG00000199217 |
. |
31489 |
0.15 |
| chr16_3915887_3916137 | 0.13 |
CREBBP |
CREB binding protein |
14109 |
0.21 |
| chr3_183296518_183296669 | 0.13 |
KLHL6 |
kelch-like family member 6 |
23116 |
0.15 |
| chr10_102242827_102242978 | 0.13 |
WNT8B |
wingless-type MMTV integration site family, member 8B |
20104 |
0.15 |
| chr11_44248571_44248722 | 0.13 |
ALX4 |
ALX homeobox 4 |
83070 |
0.09 |
| chr1_27943747_27943898 | 0.13 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
6751 |
0.15 |
| chr2_182029213_182029364 | 0.12 |
ENSG00000266705 |
. |
141091 |
0.05 |
| chr14_64330060_64330276 | 0.12 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
10436 |
0.21 |
| chr1_203257611_203257822 | 0.12 |
BTG2 |
BTG family, member 2 |
16948 |
0.16 |
| chr19_40458657_40458927 | 0.12 |
PSMC4 |
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
18120 |
0.14 |
| chr6_167532699_167533025 | 0.12 |
CCR6 |
chemokine (C-C motif) receptor 6 |
3395 |
0.24 |
| chr3_114044761_114044912 | 0.12 |
ENSG00000207770 |
. |
9420 |
0.18 |
| chr8_130987860_130988109 | 0.12 |
FAM49B |
family with sequence similarity 49, member B |
4748 |
0.22 |
| chr10_14601089_14601359 | 0.12 |
FAM107B |
family with sequence similarity 107, member B |
3045 |
0.33 |
| chr22_47080529_47080680 | 0.12 |
GRAMD4 |
GRAM domain containing 4 |
10099 |
0.22 |
| chr2_234177088_234177288 | 0.12 |
ENSG00000252010 |
. |
7185 |
0.15 |
| chr17_71737008_71737211 | 0.11 |
SDK2 |
sidekick cell adhesion molecule 2 |
96881 |
0.08 |
| chr10_11269625_11270026 | 0.11 |
RP3-323N1.2 |
|
56486 |
0.13 |
| chr18_5310391_5310755 | 0.11 |
ZBTB14 |
zinc finger and BTB domain containing 14 |
13521 |
0.26 |
| chr1_111436407_111436686 | 0.11 |
CD53 |
CD53 molecule |
20770 |
0.16 |
| chr11_59923299_59923450 | 0.11 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
25765 |
0.17 |
| chr1_229762263_229762506 | 0.11 |
URB2 |
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
403 |
0.6 |
| chr1_154302822_154303147 | 0.11 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
1709 |
0.2 |
| chr20_43520651_43520802 | 0.11 |
YWHAB |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta |
6333 |
0.15 |
| chr16_2204860_2205013 | 0.11 |
ENSG00000206630 |
. |
170 |
0.78 |
| chr1_24846109_24846267 | 0.11 |
RCAN3 |
RCAN family member 3 |
5325 |
0.18 |
| chr14_61814853_61815190 | 0.11 |
PRKCH |
protein kinase C, eta |
412 |
0.87 |
| chr2_231201045_231201221 | 0.11 |
SP140L |
SP140 nuclear body protein-like |
5652 |
0.26 |
| chr10_75817936_75818151 | 0.11 |
VCL |
vinculin |
25191 |
0.17 |
| chr7_78901447_78901598 | 0.11 |
ENSG00000212482 |
. |
71166 |
0.13 |
| chr3_43430365_43430807 | 0.11 |
SNRK-AS1 |
SNRK antisense RNA 1 |
37132 |
0.15 |
| chr19_18429556_18429758 | 0.11 |
LSM4 |
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
2588 |
0.15 |
| chr6_87897937_87898088 | 0.11 |
ZNF292 |
zinc finger protein 292 |
32673 |
0.16 |
| chr14_65457605_65457898 | 0.11 |
FNTB |
farnesyltransferase, CAAX box, beta |
4313 |
0.14 |
| chr20_39510632_39510883 | 0.10 |
ENSG00000238908 |
. |
27888 |
0.24 |
| chr20_46364251_46364402 | 0.10 |
SULF2 |
sulfatase 2 |
49907 |
0.14 |
| chr3_15133293_15133444 | 0.10 |
ZFYVE20 |
zinc finger, FYVE domain containing 20 |
7275 |
0.18 |
| chr2_231185832_231185996 | 0.10 |
SP140L |
SP140 nuclear body protein-like |
5985 |
0.26 |
| chr11_35063654_35064206 | 0.10 |
PDHX |
pyruvate dehydrogenase complex, component X |
64599 |
0.1 |
| chrX_128902828_128903127 | 0.10 |
SASH3 |
SAM and SH3 domain containing 3 |
10983 |
0.19 |
| chr2_136809758_136810032 | 0.10 |
AC093391.2 |
|
39860 |
0.17 |
| chrX_109522408_109522577 | 0.10 |
AMMECR1-IT1 |
AMMECR1 intronic transcript 1 (non-protein coding) |
27957 |
0.16 |
| chr20_419708_419969 | 0.10 |
ENSG00000206797 |
. |
11469 |
0.15 |
| chr4_6927384_6927590 | 0.10 |
TBC1D14 |
TBC1 domain family, member 14 |
15512 |
0.16 |
| chr1_206734149_206734498 | 0.10 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
3830 |
0.2 |
| chr6_91121876_91122034 | 0.10 |
ENSG00000252676 |
. |
34536 |
0.21 |
| chr2_42523029_42523463 | 0.10 |
EML4 |
echinoderm microtubule associated protein like 4 |
5147 |
0.28 |
| chr8_126304861_126305012 | 0.10 |
ENSG00000242170 |
. |
22090 |
0.22 |
| chr10_3777659_3777810 | 0.10 |
RP11-184A2.3 |
|
15525 |
0.22 |
| chr7_64461641_64461792 | 0.10 |
ERV3-1 |
endogenous retrovirus group 3, member 1 |
5315 |
0.17 |
| chr6_11439781_11439932 | 0.10 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
57275 |
0.13 |
| chr2_174833582_174833733 | 0.10 |
SP3 |
Sp3 transcription factor |
3227 |
0.39 |
| chr13_114915217_114915368 | 0.10 |
RASA3 |
RAS p21 protein activator 3 |
17206 |
0.2 |
| chr11_116798054_116798243 | 0.10 |
SIK3 |
SIK family kinase 3 |
29612 |
0.13 |
| chr9_6915201_6915352 | 0.10 |
KDM4C |
lysine (K)-specific demethylase 4C |
10225 |
0.24 |
| chr2_42425183_42425334 | 0.10 |
AC083949.1 |
|
27817 |
0.19 |
| chr4_86538528_86538679 | 0.10 |
ARHGAP24 |
Rho GTPase activating protein 24 |
13297 |
0.29 |
| chr1_109185458_109185656 | 0.10 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
18120 |
0.18 |
| chr3_72457004_72457192 | 0.09 |
RYBP |
RING1 and YY1 binding protein |
38971 |
0.21 |
| chr2_136894487_136894962 | 0.09 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
18989 |
0.26 |
| chr3_177142358_177142705 | 0.09 |
ENSG00000252028 |
. |
78809 |
0.12 |
| chr2_219181462_219181800 | 0.09 |
PNKD |
paroxysmal nonkinesigenic dyskinesia |
6265 |
0.1 |
| chr4_139997269_139997476 | 0.09 |
ELF2 |
E74-like factor 2 (ets domain transcription factor) |
7972 |
0.15 |
| chr9_21159368_21159519 | 0.09 |
ENSG00000199635 |
. |
892 |
0.45 |
| chr1_158990779_158990930 | 0.09 |
IFI16 |
interferon, gamma-inducible protein 16 |
5391 |
0.21 |
| chr1_226870909_226871060 | 0.09 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
8216 |
0.21 |
| chr6_167362176_167362468 | 0.09 |
RP11-514O12.4 |
|
7290 |
0.15 |
| chr9_134110097_134110353 | 0.09 |
NUP214 |
nucleoporin 214kDa |
6544 |
0.18 |
| chr17_26876127_26876878 | 0.09 |
UNC119 |
unc-119 homolog (C. elegans) |
85 |
0.93 |
| chr20_58836437_58836588 | 0.09 |
ENSG00000207802 |
. |
47020 |
0.19 |
| chr15_101147896_101148047 | 0.09 |
ASB7 |
ankyrin repeat and SOCS box containing 7 |
5157 |
0.16 |
| chr6_14241209_14241434 | 0.09 |
ENSG00000238987 |
. |
84628 |
0.1 |
| chr5_169731249_169731487 | 0.09 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
6137 |
0.23 |
| chr7_100846741_100846892 | 0.09 |
MOGAT3 |
monoacylglycerol O-acyltransferase 3 |
2514 |
0.14 |
| chr6_53143430_53143680 | 0.09 |
ENSG00000264056 |
. |
1764 |
0.38 |
| chr11_65553168_65553319 | 0.09 |
OVOL1 |
ovo-like zinc finger 1 |
1250 |
0.26 |
| chr2_128214705_128214856 | 0.09 |
AC010976.2 |
|
6438 |
0.15 |
| chr16_50728117_50728492 | 0.09 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
767 |
0.52 |
| chr3_151909899_151910427 | 0.09 |
MBNL1 |
muscleblind-like splicing regulator 1 |
75666 |
0.1 |
| chr19_16478953_16479699 | 0.09 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
6562 |
0.16 |
| chr16_48655371_48655896 | 0.09 |
N4BP1 |
NEDD4 binding protein 1 |
11513 |
0.18 |
| chr4_154391718_154392045 | 0.09 |
KIAA0922 |
KIAA0922 |
4380 |
0.29 |
| chr1_226873293_226873602 | 0.09 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
10679 |
0.2 |
| chr16_74619935_74620086 | 0.09 |
GLG1 |
golgi glycoprotein 1 |
20982 |
0.2 |
| chr6_30475738_30475914 | 0.09 |
XXbac-BPG249D20.9 |
|
8217 |
0.13 |
| chr7_141617224_141617397 | 0.09 |
OR9A4 |
olfactory receptor, family 9, subfamily A, member 4 |
1307 |
0.3 |
| chr20_39148848_39148999 | 0.09 |
ENSG00000252434 |
. |
103408 |
0.08 |
| chr1_8594076_8594346 | 0.09 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
8125 |
0.23 |
| chr12_92949909_92950139 | 0.09 |
ENSG00000238865 |
. |
11261 |
0.26 |
| chr1_203260343_203260644 | 0.09 |
BTG2 |
BTG family, member 2 |
14171 |
0.17 |
| chr4_40204107_40204407 | 0.09 |
RHOH |
ras homolog family member H |
2293 |
0.33 |
| chr22_31725192_31725343 | 0.09 |
RP3-400N23.6 |
|
6068 |
0.12 |
| chr5_53902252_53902403 | 0.09 |
SNX18 |
sorting nexin 18 |
88734 |
0.09 |
| chr13_24827210_24827496 | 0.09 |
SPATA13-AS1 |
SPATA13 antisense RNA 1 |
1224 |
0.37 |
| chr19_36443325_36443510 | 0.09 |
LRFN3 |
leucine rich repeat and fibronectin type III domain containing 3 |
15395 |
0.09 |
| chr5_39146620_39146886 | 0.08 |
AC008964.1 |
|
41395 |
0.17 |
| chr6_35559184_35559464 | 0.08 |
ENSG00000212579 |
. |
60271 |
0.09 |
| chr2_113937367_113937585 | 0.08 |
AC016683.5 |
|
4539 |
0.16 |
| chr15_66432505_66432656 | 0.08 |
ENSG00000263512 |
. |
100009 |
0.06 |
| chr1_153487121_153487272 | 0.08 |
ENSG00000263841 |
. |
13956 |
0.08 |
| chr18_2950342_2950493 | 0.08 |
RP11-737O24.2 |
|
2078 |
0.26 |
| chr1_226871894_226872507 | 0.08 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
9432 |
0.21 |
| chr9_95811540_95811691 | 0.08 |
SUSD3 |
sushi domain containing 3 |
9374 |
0.18 |
| chr3_32497870_32498021 | 0.08 |
CMTM6 |
CKLF-like MARVEL transmembrane domain containing 6 |
46955 |
0.14 |
| chr6_41675311_41675569 | 0.08 |
TFEB |
transcription factor EB |
1762 |
0.25 |
| chrX_41200911_41201062 | 0.08 |
ENSG00000264573 |
. |
4143 |
0.2 |
| chr5_64046210_64046361 | 0.08 |
SREK1IP1 |
SREK1-interacting protein 1 |
18227 |
0.2 |
| chr7_75029353_75029504 | 0.08 |
TRIM73 |
tripartite motif containing 73 |
1229 |
0.29 |
| chr11_64541680_64542034 | 0.08 |
SF1 |
splicing factor 1 |
3375 |
0.15 |
| chr1_154757692_154757865 | 0.08 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
74541 |
0.07 |
| chr6_24857746_24858061 | 0.08 |
ENSG00000263391 |
. |
17610 |
0.17 |
| chr22_47463732_47463883 | 0.08 |
ENSG00000221672 |
. |
220004 |
0.02 |
| chr15_77779927_77780078 | 0.08 |
HMG20A |
high mobility group 20A |
9367 |
0.26 |
| chr12_92776883_92777034 | 0.08 |
RP11-693J15.4 |
|
38349 |
0.15 |
| chr13_20539466_20539748 | 0.08 |
ZMYM2 |
zinc finger, MYM-type 2 |
6684 |
0.22 |
| chr16_17440812_17440963 | 0.08 |
XYLT1 |
xylosyltransferase I |
123851 |
0.06 |
| chr7_72435345_72435555 | 0.08 |
TRIM74 |
tripartite motif containing 74 |
2034 |
0.18 |
| chr3_13054781_13055070 | 0.08 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
26389 |
0.23 |
| chr12_46611375_46611659 | 0.08 |
SLC38A1 |
solute carrier family 38, member 1 |
49967 |
0.18 |
| chr22_42689321_42689472 | 0.08 |
TCF20 |
transcription factor 20 (AR1) |
50226 |
0.13 |
| chr3_71924513_71924711 | 0.08 |
ENSG00000239250 |
. |
48284 |
0.16 |
| chr22_42239598_42239749 | 0.08 |
RP5-821D11.7 |
|
9004 |
0.12 |
| chr3_151926787_151927119 | 0.08 |
MBNL1 |
muscleblind-like splicing regulator 1 |
58876 |
0.13 |
| chr6_139493708_139493877 | 0.08 |
HECA |
headcase homolog (Drosophila) |
37543 |
0.17 |
| chr17_40424777_40425014 | 0.08 |
AC003104.1 |
|
194 |
0.9 |
| chr4_109081069_109081352 | 0.08 |
LEF1 |
lymphoid enhancer-binding factor 1 |
6247 |
0.24 |
| chr16_3118727_3118954 | 0.08 |
ENSG00000252561 |
. |
684 |
0.37 |
| chr12_746864_747239 | 0.08 |
RP11-218M22.1 |
|
4789 |
0.18 |
| chr17_56070062_56070285 | 0.08 |
VEZF1 |
vascular endothelial zinc finger 1 |
4553 |
0.17 |
| chr17_2696074_2696428 | 0.08 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
3481 |
0.22 |
| chr1_225656318_225656532 | 0.08 |
RP11-496N12.6 |
|
3380 |
0.27 |
| chr14_22998856_22999007 | 0.08 |
TRAJ15 |
T cell receptor alpha joining 15 |
351 |
0.79 |
| chr1_113170618_113170793 | 0.08 |
ENSG00000252750 |
. |
6701 |
0.13 |
| chr12_65061545_65061862 | 0.08 |
RP11-338E21.3 |
|
12904 |
0.13 |
| chr7_104985093_104985329 | 0.08 |
ENSG00000201179 |
. |
11974 |
0.21 |
| chr5_1497877_1498028 | 0.08 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
26140 |
0.19 |
| chr1_154440668_154440819 | 0.08 |
RP11-350G8.9 |
|
11745 |
0.12 |
| chr17_8844881_8845564 | 0.08 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
23802 |
0.21 |
| chr6_133073763_133073914 | 0.08 |
RP1-55C23.7 |
|
24 |
0.96 |
| chr8_129670430_129670618 | 0.08 |
ENSG00000221351 |
. |
161516 |
0.04 |
| chr1_1542024_1542542 | 0.08 |
C1orf233 |
chromosome 1 open reading frame 233 |
6807 |
0.09 |
| chr3_56705961_56706112 | 0.08 |
FAM208A |
family with sequence similarity 208, member A |
7677 |
0.29 |
| chr15_83419735_83419886 | 0.08 |
RP11-752G15.6 |
|
474 |
0.7 |
| chr15_77300295_77300505 | 0.08 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
7752 |
0.2 |
| chr17_80517753_80517904 | 0.08 |
FOXK2 |
forkhead box K2 |
588 |
0.61 |
| chr8_29683682_29683878 | 0.08 |
ENSG00000221003 |
. |
102341 |
0.07 |
| chr22_39369446_39369597 | 0.08 |
APOBEC3B |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B |
8831 |
0.12 |
| chr13_52397484_52397635 | 0.08 |
RP11-327P2.5 |
|
19126 |
0.17 |
| chr16_84409256_84409407 | 0.08 |
ATP2C2 |
ATPase, Ca++ transporting, type 2C, member 2 |
7198 |
0.21 |
| chr16_67728613_67728764 | 0.08 |
GFOD2 |
glucose-fructose oxidoreductase domain containing 2 |
24636 |
0.08 |
| chr13_41201246_41201477 | 0.08 |
FOXO1 |
forkhead box O1 |
39373 |
0.17 |
| chr2_181911925_181912076 | 0.08 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
65250 |
0.13 |
| chr1_117311065_117311639 | 0.08 |
CD2 |
CD2 molecule |
14263 |
0.21 |
| chr5_52096687_52096895 | 0.07 |
CTD-2288O8.1 |
|
12931 |
0.19 |
| chr7_8169902_8170053 | 0.07 |
AC006042.6 |
|
16322 |
0.2 |
| chr10_14594956_14595285 | 0.07 |
FAM107B |
family with sequence similarity 107, member B |
1108 |
0.6 |
| chr6_497453_497604 | 0.07 |
RP1-20B11.2 |
|
26643 |
0.24 |
| chr17_7154048_7154378 | 0.07 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
392 |
0.45 |
| chr12_50156674_50156825 | 0.07 |
TMBIM6 |
transmembrane BAX inhibitor motif containing 6 |
10002 |
0.16 |
| chrX_123345247_123345413 | 0.07 |
ENSG00000252693 |
. |
13737 |
0.27 |
| chr4_55620528_55620679 | 0.07 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
96518 |
0.09 |
| chr9_92113326_92114071 | 0.07 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
653 |
0.53 |
| chr20_2273870_2274021 | 0.07 |
TGM3 |
transglutaminase 3 |
2702 |
0.34 |
| chr9_515997_516400 | 0.07 |
ENSG00000264949 |
. |
8645 |
0.14 |
| chr5_55909423_55909817 | 0.07 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
7561 |
0.26 |
| chr6_35703353_35703635 | 0.07 |
RP3-510O8.4 |
|
1230 |
0.28 |
| chr10_74053915_74054066 | 0.07 |
RP11-442H21.2 |
|
18252 |
0.15 |
| chr3_171862867_171863018 | 0.07 |
FNDC3B |
fibronectin type III domain containing 3B |
18178 |
0.25 |
| chr1_200976988_200977172 | 0.07 |
KIF21B |
kinesin family member 21B |
15456 |
0.18 |
| chr20_62535397_62535572 | 0.07 |
ENSG00000199805 |
. |
4824 |
0.08 |
| chr2_97010416_97010595 | 0.07 |
NCAPH |
non-SMC condensin I complex, subunit H |
8949 |
0.15 |
| chr12_58176126_58176280 | 0.07 |
TSFM |
Ts translation elongation factor, mitochondrial |
169 |
0.85 |
| chr7_37489254_37489473 | 0.07 |
ELMO1 |
engulfment and cell motility 1 |
511 |
0.8 |
| chrX_15815885_15816036 | 0.07 |
ZRSR2 |
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
7365 |
0.21 |
| chr11_77159328_77159479 | 0.07 |
DKFZP434E1119 |
|
25013 |
0.18 |
| chr22_27071985_27072283 | 0.07 |
CRYBA4 |
crystallin, beta A4 |
54206 |
0.12 |
| chr11_67046455_67046752 | 0.07 |
ANKRD13D |
ankyrin repeat domain 13 family, member D |
9415 |
0.11 |
| chr3_182683857_182684049 | 0.07 |
DCUN1D1 |
DCN1, defective in cullin neddylation 1, domain containing 1 |
13986 |
0.2 |
| chr3_107789495_107789646 | 0.07 |
CD47 |
CD47 molecule |
11191 |
0.3 |
| chr7_155131318_155131469 | 0.07 |
BLACE |
B-cell acute lymphoblastic leukemia expressed |
29236 |
0.15 |
| chr1_165819042_165819193 | 0.07 |
UCK2 |
uridine-cytidine kinase 2 |
22058 |
0.17 |
| chrX_135669720_135670141 | 0.07 |
ENSG00000252320 |
. |
4531 |
0.17 |
| chr12_29318585_29318886 | 0.07 |
FAR2 |
fatty acyl CoA reductase 2 |
16602 |
0.24 |
| chr1_43273074_43273425 | 0.07 |
CCDC23 |
coiled-coil domain containing 23 |
9226 |
0.12 |
| chrX_19729925_19730220 | 0.07 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
28883 |
0.25 |
| chr10_116299368_116299596 | 0.07 |
ABLIM1 |
actin binding LIM protein 1 |
12792 |
0.27 |
| chr2_237878465_237879184 | 0.07 |
ENSG00000202341 |
. |
45124 |
0.17 |
| chr21_36318885_36319060 | 0.07 |
RUNX1 |
runt-related transcription factor 1 |
56885 |
0.17 |
| chr9_110377144_110377295 | 0.07 |
ENSG00000202308 |
. |
48308 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |