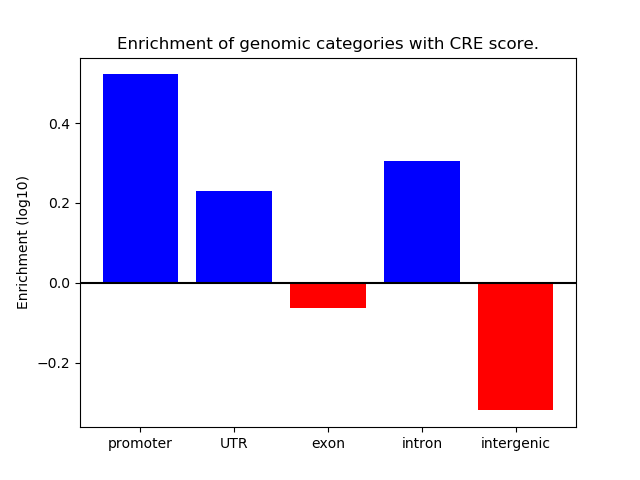
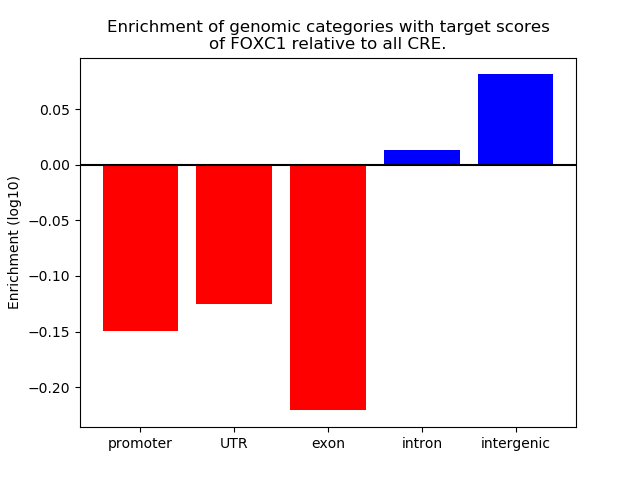
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
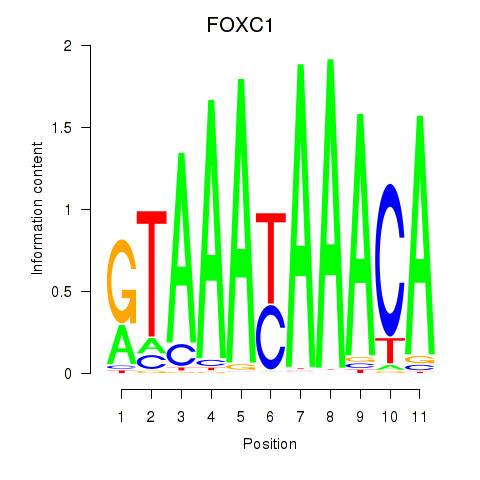
Results for FOXC1
Z-value: 1.37
Transcription factors associated with FOXC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXC1
|
ENSG00000054598.5 | forkhead box C1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_1703338_1703561 | FOXC1 | 92768 | 0.094534 | 0.87 | 2.5e-03 | Click! |
| chr6_1575832_1575983 | FOXC1 | 34774 | 0.209341 | 0.86 | 3.2e-03 | Click! |
| chr6_1686574_1686725 | FOXC1 | 75968 | 0.121959 | 0.85 | 3.8e-03 | Click! |
| chr6_1772728_1772879 | FOXC1 | 162122 | 0.039404 | -0.81 | 7.7e-03 | Click! |
| chr6_1698536_1698687 | FOXC1 | 87930 | 0.101592 | 0.80 | 8.9e-03 | Click! |
Activity of the FOXC1 motif across conditions
Conditions sorted by the z-value of the FOXC1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
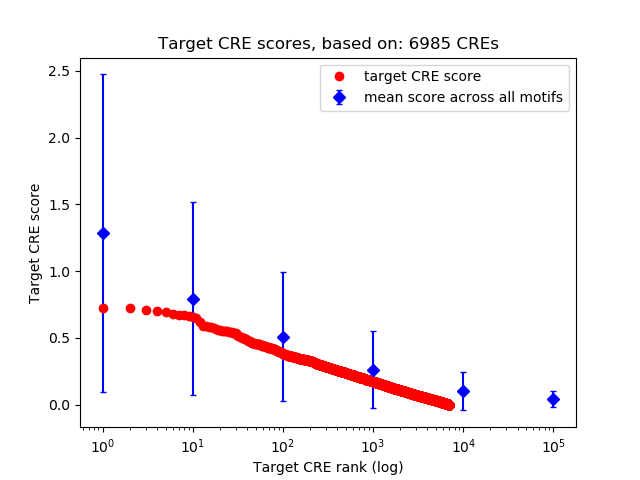
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_157242892_157243172 | 0.73 |
ENSG00000221189 |
. |
25234 |
0.27 |
| chr10_95507444_95507712 | 0.72 |
LGI1 |
leucine-rich, glioma inactivated 1 |
9988 |
0.23 |
| chr19_18575570_18575881 | 0.71 |
ELL |
elongation factor RNA polymerase II |
11018 |
0.1 |
| chr1_40358181_40358689 | 0.70 |
RP1-118J21.5 |
|
4982 |
0.14 |
| chr20_36904040_36904191 | 0.69 |
KIAA1755 |
KIAA1755 |
14941 |
0.15 |
| chr20_47448190_47448648 | 0.68 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
3999 |
0.32 |
| chr2_12959099_12960371 | 0.67 |
ENSG00000264370 |
. |
82242 |
0.11 |
| chr8_126536876_126537027 | 0.67 |
ENSG00000266452 |
. |
80144 |
0.1 |
| chr18_61497949_61498456 | 0.66 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
40724 |
0.14 |
| chr18_457195_458136 | 0.66 |
RP11-720L2.2 |
|
33249 |
0.16 |
| chr12_66050940_66051359 | 0.65 |
HMGA2 |
high mobility group AT-hook 2 |
166762 |
0.03 |
| chr3_119921549_119921940 | 0.62 |
ENSG00000206721 |
. |
7423 |
0.22 |
| chr1_115876554_115876760 | 0.59 |
NGF |
nerve growth factor (beta polypeptide) |
4200 |
0.3 |
| chr1_205144955_205145106 | 0.59 |
DSTYK |
dual serine/threonine and tyrosine protein kinase |
35664 |
0.13 |
| chr10_115910998_115911149 | 0.58 |
C10orf118 |
chromosome 10 open reading frame 118 |
6712 |
0.18 |
| chr4_103583867_103584018 | 0.58 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
84921 |
0.08 |
| chr13_44811098_44811768 | 0.58 |
SMIM2 |
small integral membrane protein 2 |
76040 |
0.1 |
| chr12_65700883_65701407 | 0.57 |
MSRB3 |
methionine sulfoxide reductase B3 |
19510 |
0.22 |
| chr5_121462821_121463158 | 0.56 |
ZNF474 |
zinc finger protein 474 |
2219 |
0.3 |
| chr8_131377178_131377403 | 0.56 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
21833 |
0.28 |
| chr4_157700822_157700973 | 0.55 |
RP11-154F14.2 |
|
61614 |
0.13 |
| chr5_77797186_77797364 | 0.55 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
47699 |
0.18 |
| chr19_6745452_6745603 | 0.55 |
TRIP10 |
thyroid hormone receptor interactor 10 |
4598 |
0.13 |
| chr1_159852066_159852217 | 0.55 |
RP11-190A12.8 |
|
15397 |
0.08 |
| chr11_36178088_36178239 | 0.55 |
COMMD9 |
COMM domain containing 9 |
132811 |
0.05 |
| chr1_225942912_225943116 | 0.54 |
ENSG00000223306 |
. |
14037 |
0.14 |
| chr5_65169471_65169622 | 0.54 |
ERBB2IP |
erbb2 interacting protein |
52757 |
0.14 |
| chr20_52235971_52236462 | 0.54 |
ZNF217 |
zinc finger protein 217 |
10785 |
0.21 |
| chr2_29175143_29175407 | 0.54 |
FAM179A |
family with sequence similarity 179, member A |
4202 |
0.14 |
| chr11_57267439_57267663 | 0.53 |
SLC43A1 |
solute carrier family 43 (amino acid system L transporter), member 1 |
1241 |
0.29 |
| chr16_56944713_56945265 | 0.52 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
20971 |
0.1 |
| chr7_79780258_79780623 | 0.52 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
15369 |
0.27 |
| chr19_41768990_41769258 | 0.51 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
548 |
0.66 |
| chr6_110868489_110868770 | 0.51 |
CTA-331P3.1 |
|
69519 |
0.1 |
| chr2_43242041_43242192 | 0.50 |
ENSG00000207087 |
. |
76516 |
0.11 |
| chr7_80261706_80261971 | 0.50 |
CD36 |
CD36 molecule (thrombospondin receptor) |
6122 |
0.33 |
| chr10_88891932_88892287 | 0.50 |
RP11-96C23.9 |
|
18398 |
0.13 |
| chr3_54925868_54926132 | 0.49 |
CACNA2D3-AS1 |
CACNA2D3 antisense RNA 1 |
9282 |
0.27 |
| chr11_110634298_110634449 | 0.49 |
ARHGAP20 |
Rho GTPase activating protein 20 |
50461 |
0.19 |
| chr13_45769539_45769983 | 0.48 |
KCTD4 |
potassium channel tetramerization domain containing 4 |
909 |
0.6 |
| chr1_98403274_98403477 | 0.48 |
DPYD |
dihydropyrimidine dehydrogenase |
16770 |
0.28 |
| chr11_15919049_15919200 | 0.48 |
CTD-3096P4.1 |
|
125612 |
0.06 |
| chr10_18073243_18073394 | 0.47 |
MRC1 |
mannose receptor, C type 1 |
25034 |
0.17 |
| chr15_67803044_67803195 | 0.47 |
C15orf61 |
chromosome 15 open reading frame 61 |
10287 |
0.21 |
| chr10_75646701_75646885 | 0.47 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
12450 |
0.12 |
| chr8_119756248_119756399 | 0.46 |
SAMD12 |
sterile alpha motif domain containing 12 |
122089 |
0.06 |
| chr1_43760092_43760243 | 0.46 |
TIE1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
6497 |
0.13 |
| chr6_16676788_16676939 | 0.46 |
RP1-151F17.1 |
|
84506 |
0.1 |
| chr1_6930430_6930581 | 0.46 |
CAMTA1 |
calmodulin binding transcription activator 1 |
84914 |
0.08 |
| chr12_116814280_116814431 | 0.46 |
ENSG00000264037 |
. |
51768 |
0.15 |
| chr3_58374430_58375078 | 0.46 |
PXK |
PX domain containing serine/threonine kinase |
6647 |
0.2 |
| chr18_20960634_20960918 | 0.45 |
TMEM241 |
transmembrane protein 241 |
57041 |
0.11 |
| chr10_17826265_17826416 | 0.45 |
MRC1L1 |
cDNA FLJ56855, highly similar to Macrophage mannose receptor 1 |
25022 |
0.16 |
| chr9_94908980_94909563 | 0.45 |
ENSG00000238996 |
. |
1803 |
0.38 |
| chr8_96111972_96112794 | 0.45 |
RP11-320N21.2 |
|
12231 |
0.16 |
| chr18_22907594_22907986 | 0.45 |
ZNF521 |
zinc finger protein 521 |
23367 |
0.23 |
| chr1_243428429_243428713 | 0.45 |
SDCCAG8 |
serologically defined colon cancer antigen 8 |
9199 |
0.19 |
| chr13_41639007_41639306 | 0.44 |
WBP4 |
WW domain binding protein 4 |
3460 |
0.19 |
| chr11_5627634_5627785 | 0.44 |
TRIM6 |
tripartite motif containing 6 |
9673 |
0.1 |
| chr18_60905822_60905974 | 0.44 |
ENSG00000238988 |
. |
44000 |
0.14 |
| chr5_88851937_88852088 | 0.44 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
137388 |
0.05 |
| chr9_117439245_117439536 | 0.44 |
ENSG00000272241 |
. |
60563 |
0.1 |
| chr12_46796862_46797190 | 0.44 |
SLC38A2 |
solute carrier family 38, member 2 |
30376 |
0.2 |
| chr1_98315176_98315780 | 0.44 |
DPYD-AS2 |
DPYD antisense RNA 2 |
52957 |
0.16 |
| chr3_188390892_188391164 | 0.44 |
ENSG00000207651 |
. |
15541 |
0.22 |
| chr22_30578717_30578868 | 0.43 |
RP3-438O4.4 |
|
24306 |
0.13 |
| chr11_34449252_34449782 | 0.43 |
CAT |
catalase |
10955 |
0.23 |
| chr20_5627905_5628088 | 0.43 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
36324 |
0.2 |
| chr11_100720535_100720686 | 0.43 |
ENSG00000200047 |
. |
10249 |
0.23 |
| chr9_86126228_86126614 | 0.43 |
FRMD3 |
FERM domain containing 3 |
26932 |
0.16 |
| chr16_11143336_11143487 | 0.43 |
RP11-66H6.3 |
|
21548 |
0.18 |
| chr5_93553629_93553780 | 0.42 |
FAM172A |
family with sequence similarity 172, member A |
106323 |
0.08 |
| chr5_133927870_133928021 | 0.42 |
SAR1B |
SAR1 homolog B (S. cerevisiae) |
20831 |
0.15 |
| chr2_72450802_72450953 | 0.42 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
75710 |
0.13 |
| chr6_164530035_164530217 | 0.42 |
ENSG00000266128 |
. |
268533 |
0.02 |
| chrX_136134760_136135069 | 0.42 |
GPR101 |
G protein-coupled receptor 101 |
21081 |
0.24 |
| chr5_115789878_115790274 | 0.42 |
CTB-118N6.3 |
|
4650 |
0.26 |
| chr7_23512466_23512617 | 0.42 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
2455 |
0.25 |
| chr2_75052279_75052565 | 0.42 |
HK2 |
hexokinase 2 |
8686 |
0.26 |
| chr4_148694501_148694652 | 0.41 |
ENSG00000264274 |
. |
9170 |
0.18 |
| chr15_77695649_77695800 | 0.41 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
16718 |
0.22 |
| chr6_37028055_37028341 | 0.41 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
15591 |
0.19 |
| chr21_46550357_46550680 | 0.41 |
ADARB1 |
adenosine deaminase, RNA-specific, B1 |
9703 |
0.16 |
| chr6_2631111_2631262 | 0.41 |
C6orf195 |
chromosome 6 open reading frame 195 |
3651 |
0.34 |
| chr2_144711401_144711552 | 0.40 |
AC016910.1 |
|
16836 |
0.25 |
| chr3_78694842_78694993 | 0.40 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
11187 |
0.32 |
| chr3_37156478_37156973 | 0.40 |
ENSG00000206645 |
. |
22000 |
0.14 |
| chr14_78060038_78060870 | 0.40 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
3210 |
0.25 |
| chr10_105595246_105595397 | 0.40 |
SH3PXD2A |
SH3 and PX domains 2A |
19843 |
0.19 |
| chr15_100584069_100584220 | 0.40 |
ENSG00000252957 |
. |
30251 |
0.17 |
| chr11_86884640_86884791 | 0.40 |
TMEM135 |
transmembrane protein 135 |
135633 |
0.05 |
| chr2_226983007_226983256 | 0.39 |
ENSG00000263363 |
. |
540378 |
0.0 |
| chr8_19045527_19045678 | 0.39 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
103362 |
0.08 |
| chr11_86355699_86355850 | 0.39 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
27466 |
0.25 |
| chr6_72188929_72189086 | 0.39 |
ENSG00000212099 |
. |
59672 |
0.12 |
| chr7_41909334_41909592 | 0.39 |
AC005027.3 |
|
164530 |
0.04 |
| chr4_40623166_40623743 | 0.39 |
RBM47 |
RNA binding motif protein 47 |
8427 |
0.27 |
| chr15_59461940_59462091 | 0.38 |
ENSG00000253030 |
. |
1446 |
0.28 |
| chr8_119427422_119427697 | 0.38 |
AC023590.1 |
Uncharacterized protein |
133078 |
0.05 |
| chr16_70779927_70780078 | 0.38 |
RP11-394B2.6 |
|
958 |
0.42 |
| chr14_68454710_68454913 | 0.38 |
CTD-2566J3.1 |
|
142102 |
0.04 |
| chr6_56406545_56407042 | 0.38 |
DST |
dystonin |
68005 |
0.13 |
| chr12_29895057_29895338 | 0.38 |
TMTC1 |
transmembrane and tetratricopeptide repeat containing 1 |
41534 |
0.21 |
| chr4_22515546_22516022 | 0.38 |
GPR125 |
G protein-coupled receptor 125 |
1836 |
0.52 |
| chr1_234630412_234630563 | 0.38 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
15638 |
0.21 |
| chr2_13118868_13119112 | 0.38 |
ENSG00000264370 |
. |
241497 |
0.02 |
| chr1_64292995_64293146 | 0.38 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
53356 |
0.16 |
| chr11_69084536_69085027 | 0.37 |
MYEOV |
myeloma overexpressed |
23156 |
0.25 |
| chr11_44257214_44257365 | 0.37 |
ALX4 |
ALX homeobox 4 |
74427 |
0.11 |
| chr12_52182216_52182367 | 0.37 |
AC068987.1 |
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
21498 |
0.2 |
| chr11_126788047_126788198 | 0.37 |
KIRREL3-AS2 |
KIRREL3 antisense RNA 2 |
22520 |
0.2 |
| chr9_18638757_18638908 | 0.37 |
ENSG00000252960 |
. |
12313 |
0.27 |
| chr6_112366386_112366743 | 0.37 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
8711 |
0.21 |
| chr19_56059176_56059327 | 0.37 |
SBK3 |
SH3 domain binding kinase family, member 3 |
2342 |
0.14 |
| chr3_11278939_11279235 | 0.37 |
HRH1 |
histamine receptor H1 |
11370 |
0.25 |
| chr16_17266652_17267123 | 0.37 |
CTD-2576D5.4 |
|
38526 |
0.23 |
| chr13_37018901_37019079 | 0.37 |
CCNA1 |
cyclin A1 |
12495 |
0.23 |
| chr3_11279617_11279863 | 0.37 |
HRH1 |
histamine receptor H1 |
12023 |
0.25 |
| chr1_172872813_172873091 | 0.37 |
TNFSF18 |
tumor necrosis factor (ligand) superfamily, member 18 |
147104 |
0.04 |
| chr20_32922542_32922693 | 0.36 |
AHCY |
adenosylhomocysteinase |
23009 |
0.13 |
| chr5_36367141_36367292 | 0.36 |
RANBP3L |
RAN binding protein 3-like |
65000 |
0.12 |
| chr8_120553418_120553569 | 0.36 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
51755 |
0.15 |
| chr14_54360916_54361067 | 0.36 |
ENSG00000266431 |
. |
54211 |
0.16 |
| chr7_1555587_1555854 | 0.36 |
AC102953.6 |
|
6657 |
0.14 |
| chr3_149274344_149274603 | 0.36 |
WWTR1 |
WW domain containing transcription regulator 1 |
19566 |
0.18 |
| chr1_39608036_39608283 | 0.36 |
ENSG00000222378 |
. |
11809 |
0.17 |
| chr6_140053583_140053734 | 0.36 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
357901 |
0.01 |
| chr1_181047325_181047476 | 0.36 |
IER5 |
immediate early response 5 |
10238 |
0.21 |
| chr4_83992439_83992590 | 0.36 |
COPS4 |
COP9 signalosome subunit 4 |
36136 |
0.14 |
| chr13_32520415_32520566 | 0.36 |
EEF1DP3 |
eukaryotic translation elongation factor 1 delta pseudogene 3 |
6227 |
0.29 |
| chr13_111062611_111062762 | 0.36 |
ENSG00000238629 |
. |
3866 |
0.28 |
| chr2_145252336_145252641 | 0.36 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
22627 |
0.22 |
| chr2_174862628_174862955 | 0.36 |
SP3 |
Sp3 transcription factor |
32361 |
0.24 |
| chr12_46949120_46949271 | 0.36 |
SLC38A2 |
solute carrier family 38, member 2 |
182545 |
0.03 |
| chr3_114225823_114225974 | 0.36 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
52234 |
0.16 |
| chr2_216400648_216400799 | 0.36 |
AC012462.1 |
|
99747 |
0.08 |
| chr12_96866193_96866344 | 0.35 |
C12orf55 |
chromosome 12 open reading frame 55 |
17085 |
0.19 |
| chr12_57542255_57543089 | 0.35 |
RP11-545N8.3 |
|
1270 |
0.29 |
| chr1_161762804_161762955 | 0.35 |
ATF6 |
activating transcription factor 6 |
26795 |
0.12 |
| chr8_48523757_48523908 | 0.35 |
SPIDR |
scaffolding protein involved in DNA repair |
48394 |
0.14 |
| chr4_19750489_19750640 | 0.35 |
SLIT2 |
slit homolog 2 (Drosophila) |
504319 |
0.0 |
| chr9_16626415_16626566 | 0.35 |
RP11-62F24.1 |
|
816 |
0.74 |
| chr10_131926285_131926436 | 0.35 |
GLRX3 |
glutaredoxin 3 |
8303 |
0.32 |
| chr10_51573717_51573868 | 0.35 |
NCOA4 |
nuclear receptor coactivator 4 |
492 |
0.79 |
| chr8_98963963_98964114 | 0.35 |
MATN2 |
matrilin 2 |
1071 |
0.57 |
| chr9_22683470_22683621 | 0.35 |
DMRTA1 |
DMRT-like family A1 |
236705 |
0.02 |
| chr5_71426364_71426515 | 0.35 |
MAP1B |
microtubule-associated protein 1B |
23126 |
0.22 |
| chr20_52353487_52354058 | 0.35 |
ENSG00000238468 |
. |
68475 |
0.12 |
| chr20_18205406_18205557 | 0.35 |
ENSG00000266720 |
. |
34014 |
0.11 |
| chr2_20503901_20504052 | 0.35 |
PUM2 |
pumilio RNA-binding family member 2 |
8041 |
0.2 |
| chr9_124716545_124716696 | 0.35 |
RP11-244O19.1 |
|
134889 |
0.04 |
| chr6_148852882_148853033 | 0.35 |
ENSG00000223322 |
. |
7581 |
0.32 |
| chr13_37362298_37362449 | 0.35 |
RFXAP |
regulatory factor X-associated protein |
30988 |
0.17 |
| chr21_30620651_30620953 | 0.34 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
50390 |
0.1 |
| chr3_149865923_149866214 | 0.34 |
RP11-167H9.4 |
|
50249 |
0.14 |
| chr7_114563503_114563973 | 0.34 |
MDFIC |
MyoD family inhibitor domain containing |
607 |
0.85 |
| chr16_71916526_71917374 | 0.34 |
ZNF821 |
zinc finger protein 821 |
462 |
0.56 |
| chr2_44347955_44348155 | 0.34 |
ENSG00000252599 |
. |
33979 |
0.16 |
| chr19_11454879_11455424 | 0.34 |
CCDC159 |
coiled-coil domain containing 159 |
826 |
0.33 |
| chr8_62632405_62632737 | 0.34 |
ENSG00000264408 |
. |
5224 |
0.25 |
| chr14_80050698_80050849 | 0.34 |
RP11-242P2.2 |
|
48881 |
0.19 |
| chr2_206411815_206412034 | 0.34 |
NRP2 |
neuropilin 2 |
135300 |
0.05 |
| chr15_42119496_42120104 | 0.34 |
RP11-23P13.4 |
|
253 |
0.47 |
| chr6_12608393_12608818 | 0.34 |
PHACTR1 |
phosphatase and actin regulator 1 |
109288 |
0.07 |
| chr11_122009054_122009205 | 0.34 |
ENSG00000198975 |
. |
8172 |
0.16 |
| chr10_104573498_104574150 | 0.34 |
ENSG00000252994 |
. |
10138 |
0.12 |
| chr6_139868425_139868726 | 0.34 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
172818 |
0.03 |
| chr7_95064533_95064791 | 0.34 |
PON2 |
paraoxonase 2 |
152 |
0.95 |
| chr2_144854247_144854398 | 0.34 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
138982 |
0.05 |
| chr20_19795785_19795937 | 0.34 |
AL121761.2 |
Uncharacterized protein |
57182 |
0.12 |
| chr16_29547755_29547906 | 0.34 |
ENSG00000239193 |
. |
2856 |
0.19 |
| chr6_81348783_81348934 | 0.34 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
532475 |
0.0 |
| chr14_68466602_68466753 | 0.34 |
CTD-2566J3.1 |
|
130236 |
0.05 |
| chr4_54408278_54408429 | 0.34 |
LNX1 |
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
16031 |
0.2 |
| chr6_157737936_157738117 | 0.34 |
TMEM242 |
transmembrane protein 242 |
6607 |
0.24 |
| chr22_24879293_24879554 | 0.34 |
ADORA2A-AS1 |
ADORA2A antisense RNA 1 |
11340 |
0.14 |
| chr12_77626070_77626221 | 0.34 |
ENSG00000238769 |
. |
69340 |
0.14 |
| chr7_40713134_40713414 | 0.34 |
AC005160.3 |
|
101883 |
0.08 |
| chr12_91533860_91534011 | 0.34 |
DCN |
decorin |
6050 |
0.26 |
| chr16_14653487_14653730 | 0.34 |
ENSG00000242430 |
. |
42343 |
0.12 |
| chr9_136423569_136424056 | 0.34 |
FAM163B |
family with sequence similarity 163, member B |
21556 |
0.12 |
| chr11_12699502_12699653 | 0.34 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
2527 |
0.41 |
| chr4_81158071_81158222 | 0.34 |
FGF5 |
fibroblast growth factor 5 |
29607 |
0.18 |
| chr9_4144869_4145020 | 0.33 |
GLIS3 |
GLIS family zinc finger 3 |
249 |
0.95 |
| chr10_129076543_129076694 | 0.33 |
FAM196A |
family with sequence similarity 196, member A |
82196 |
0.11 |
| chr3_135794639_135794790 | 0.33 |
RP11-305O4.3 |
|
12041 |
0.25 |
| chr4_56049873_56050024 | 0.33 |
ENSG00000239545 |
. |
32131 |
0.17 |
| chr1_39678600_39678751 | 0.33 |
RP11-416A14.1 |
|
6491 |
0.18 |
| chr21_28944974_28945125 | 0.33 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
606217 |
0.0 |
| chr21_27777769_27777920 | 0.33 |
AP001597.1 |
|
418 |
0.88 |
| chr7_100782348_100782499 | 0.33 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
12044 |
0.1 |
| chr6_116685228_116685657 | 0.33 |
DSE |
dermatan sulfate epimerase |
5568 |
0.22 |
| chr3_175149023_175149174 | 0.33 |
ENSG00000199792 |
. |
33961 |
0.21 |
| chr3_99832344_99832495 | 0.33 |
FILIP1L |
filamin A interacting protein 1-like |
938 |
0.47 |
| chr17_28181951_28182102 | 0.33 |
RP11-338L22.2 |
|
8394 |
0.15 |
| chr20_10450434_10450690 | 0.33 |
SLX4IP |
SLX4 interacting protein |
34611 |
0.17 |
| chr3_188374207_188374358 | 0.33 |
ENSG00000264114 |
. |
25792 |
0.18 |
| chr2_174016293_174016444 | 0.33 |
MLK7-AS1 |
MLK7 antisense RNA 1 |
55972 |
0.13 |
| chr7_133331165_133331316 | 0.33 |
EXOC4 |
exocyst complex component 4 |
70024 |
0.14 |
| chr13_21654358_21654593 | 0.33 |
LATS2 |
large tumor suppressor kinase 2 |
18789 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 0.3 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.1 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.1 | 0.3 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.4 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.1 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.2 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.0 | GO:0072191 | positive regulation of smooth muscle cell differentiation(GO:0051152) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.3 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.0 | GO:2000043 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.0 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.4 | GO:0050922 | negative regulation of chemotaxis(GO:0050922) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:1903170 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.3 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.0 | GO:0002885 | positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.2 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.3 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.3 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.3 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.0 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0048263 | determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.1 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.4 | GO:0061641 | DNA replication-independent nucleosome assembly(GO:0006336) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0043306 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:2000757 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:0006693 | prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:1904478 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.2 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0010875 | positive regulation of cholesterol efflux(GO:0010875) |
| 0.0 | 0.4 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.0 | GO:0072124 | glomerular mesangial cell proliferation(GO:0072110) regulation of glomerular mesangial cell proliferation(GO:0072124) |
| 0.0 | 0.0 | GO:0033133 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0010611 | regulation of cardiac muscle hypertrophy(GO:0010611) |
| 0.0 | 0.0 | GO:1902624 | positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.0 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.0 | GO:0060192 | negative regulation of lipase activity(GO:0060192) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.2 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.0 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.0 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.2 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0090114 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.2 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.0 | 0.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.2 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.0 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.0 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.0 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0043256 | laminin complex(GO:0043256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.5 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.1 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.2 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.3 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.0 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.0 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |