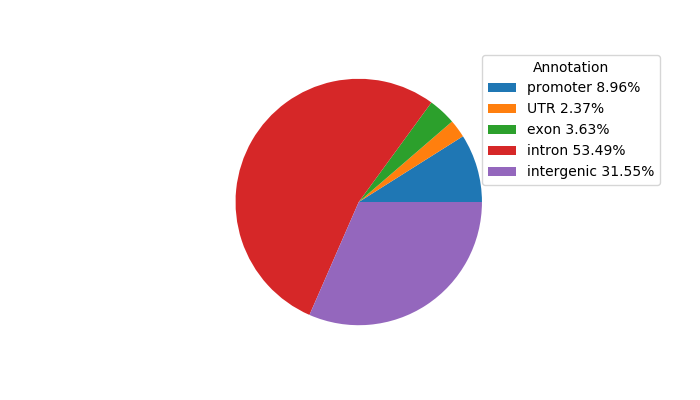
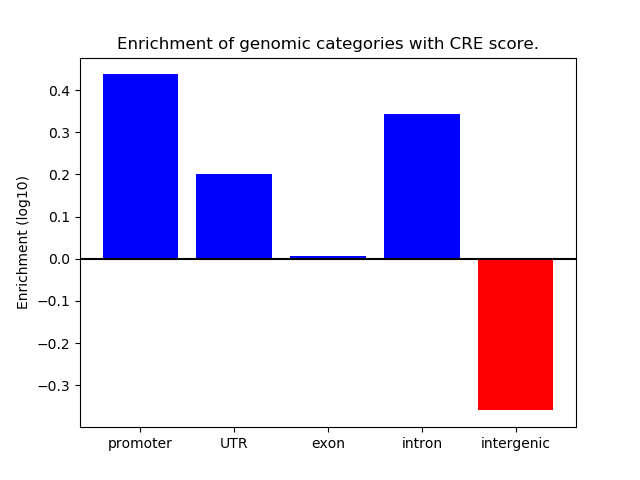
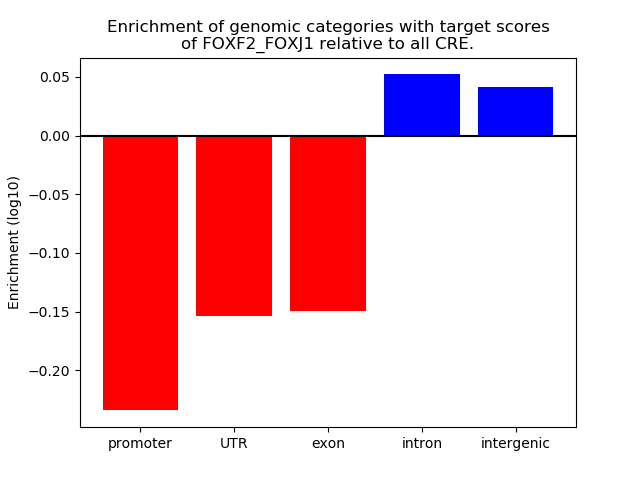
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for FOXF2_FOXJ1
Z-value: 1.09
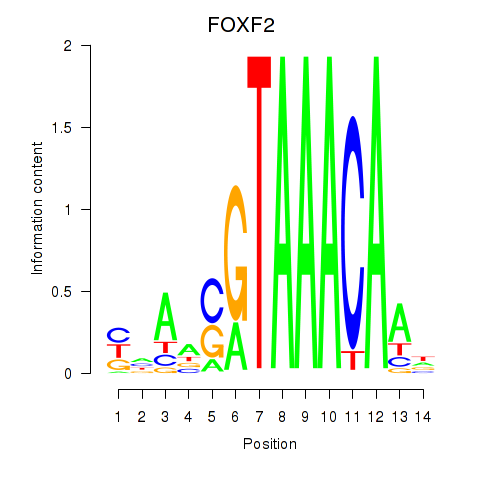
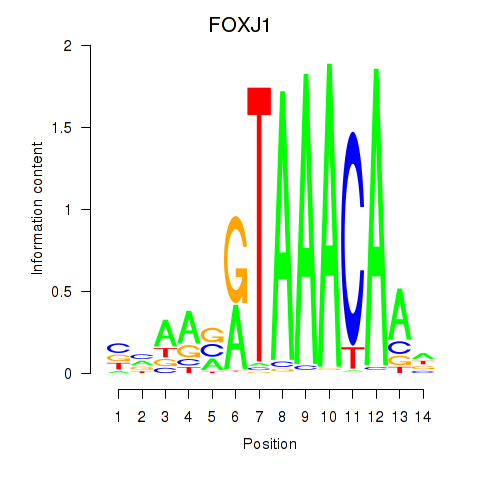
Transcription factors associated with FOXF2_FOXJ1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXF2
|
ENSG00000137273.3 | forkhead box F2 |
|
FOXJ1
|
ENSG00000129654.7 | forkhead box J1 |
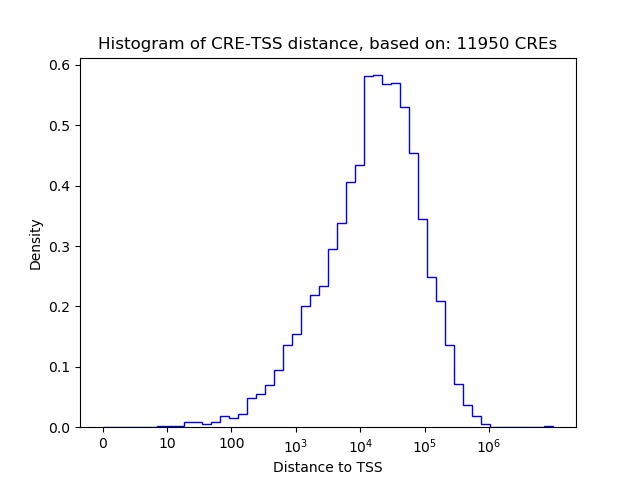
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_1407221_1407459 | FOXF2 | 17271 | 0.210329 | 0.46 | 2.1e-01 | Click! |
| chr6_1389149_1389347 | FOXF2 | 821 | 0.656661 | -0.42 | 2.6e-01 | Click! |
| chr6_1406853_1407004 | FOXF2 | 16859 | 0.211014 | 0.38 | 3.1e-01 | Click! |
| chr6_1388815_1389115 | FOXF2 | 1104 | 0.539527 | 0.35 | 3.6e-01 | Click! |
| chr6_1388553_1388704 | FOXF2 | 1441 | 0.438898 | -0.34 | 3.7e-01 | Click! |
| chr17_74137011_74137327 | FOXJ1 | 211 | 0.695528 | -0.62 | 7.7e-02 | Click! |
Activity of the FOXF2_FOXJ1 motif across conditions
Conditions sorted by the z-value of the FOXF2_FOXJ1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
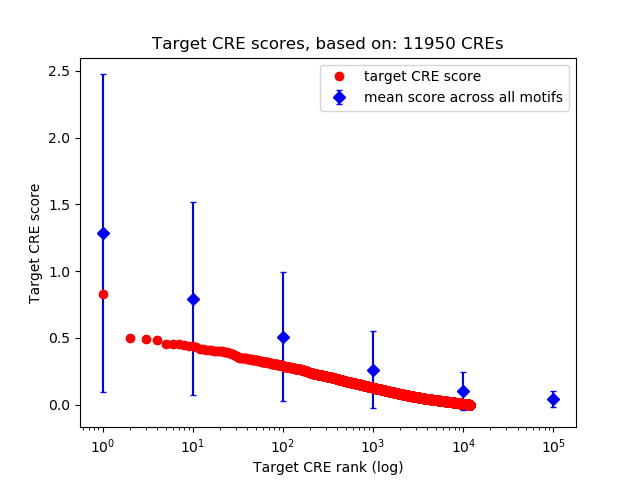
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_80354102_80354437 | 0.83 |
RP13-20L14.4 |
|
4989 |
0.1 |
| chr16_49733508_49733659 | 0.50 |
ENSG00000221134 |
. |
20452 |
0.22 |
| chr1_198859851_198860145 | 0.49 |
ENSG00000207759 |
. |
31716 |
0.2 |
| chr6_26350885_26351036 | 0.48 |
ENSG00000252399 |
. |
2304 |
0.16 |
| chr8_13089426_13089577 | 0.45 |
DLC1 |
deleted in liver cancer 1 |
44554 |
0.17 |
| chr5_148290573_148290835 | 0.45 |
RP11-44B19.1 |
|
59199 |
0.12 |
| chr10_79669456_79669735 | 0.45 |
ENSG00000243446 |
. |
2709 |
0.24 |
| chr8_19069882_19070033 | 0.45 |
SH2D4A |
SH2 domain containing 4A |
101171 |
0.08 |
| chr4_119674096_119674247 | 0.44 |
SEC24D |
SEC24 family member D |
5626 |
0.29 |
| chr7_111078523_111078674 | 0.44 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
45627 |
0.21 |
| chr4_150925367_150925518 | 0.43 |
ENSG00000207121 |
. |
69914 |
0.11 |
| chr8_108470356_108470589 | 0.42 |
ANGPT1 |
angiopoietin 1 |
36751 |
0.24 |
| chr18_60548793_60548944 | 0.42 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
21515 |
0.2 |
| chr6_56573158_56573456 | 0.41 |
DST |
dystonin |
65513 |
0.14 |
| chr1_85065842_85065993 | 0.41 |
CTBS |
chitobiase, di-N-acetyl- |
25770 |
0.17 |
| chr5_14397094_14397245 | 0.41 |
TRIO |
trio Rho guanine nucleotide exchange factor |
91388 |
0.09 |
| chr9_94539443_94539824 | 0.41 |
ENSG00000266855 |
. |
141100 |
0.05 |
| chr13_33803306_33803457 | 0.41 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
23238 |
0.2 |
| chr12_109539613_109539764 | 0.40 |
UNG |
uracil-DNA glycosylase |
3765 |
0.15 |
| chr6_166985772_166986271 | 0.40 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
27843 |
0.18 |
| chr6_158838381_158838532 | 0.40 |
RP11-732M18.2 |
|
18863 |
0.2 |
| chr18_19904044_19904557 | 0.40 |
ENSG00000238907 |
. |
63761 |
0.11 |
| chr3_64330269_64330420 | 0.40 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
76689 |
0.1 |
| chr12_66089003_66089154 | 0.39 |
HMGA2 |
high mobility group AT-hook 2 |
128833 |
0.05 |
| chr3_189434335_189434486 | 0.39 |
TP63 |
tumor protein p63 |
73039 |
0.12 |
| chr3_69469991_69470142 | 0.39 |
FRMD4B |
FERM domain containing 4B |
34636 |
0.23 |
| chr8_28984494_28984645 | 0.38 |
CTD-2647L4.1 |
|
12992 |
0.15 |
| chr7_134049333_134049484 | 0.38 |
SLC35B4 |
solute carrier family 35 (UDP-xylose/UDP-N-acetylglucosamine transporter), member B4 |
47605 |
0.16 |
| chr17_48137007_48137298 | 0.37 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
3348 |
0.18 |
| chr1_162319832_162320115 | 0.36 |
ENSG00000207729 |
. |
7637 |
0.11 |
| chr8_80672969_80673271 | 0.36 |
HEY1 |
hes-related family bHLH transcription factor with YRPW motif 1 |
5590 |
0.28 |
| chr1_12483776_12483927 | 0.36 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
13927 |
0.23 |
| chr12_43709758_43710210 | 0.35 |
ENSG00000215993 |
. |
24601 |
0.27 |
| chr12_10794215_10794366 | 0.35 |
STYK1 |
serine/threonine/tyrosine kinase 1 |
12053 |
0.16 |
| chr11_29315204_29315355 | 0.35 |
ENSG00000211499 |
. |
17043 |
0.31 |
| chr11_5310917_5311068 | 0.35 |
OR51B4 |
olfactory receptor, family 51, subfamily B, member 4 |
12234 |
0.09 |
| chr1_151592190_151592613 | 0.35 |
RP11-404E16.1 |
|
6562 |
0.11 |
| chr2_238365295_238365446 | 0.35 |
MLPH |
melanophilin |
28701 |
0.15 |
| chr3_119921549_119921940 | 0.35 |
ENSG00000206721 |
. |
7423 |
0.22 |
| chr5_102607577_102607797 | 0.34 |
C5orf30 |
chromosome 5 open reading frame 30 |
12562 |
0.23 |
| chr3_72868449_72868731 | 0.34 |
SHQ1 |
SHQ1, H/ACA ribonucleoprotein assembly factor |
26924 |
0.21 |
| chr11_9173045_9173196 | 0.34 |
DENND5A |
DENN/MADD domain containing 5A |
834 |
0.63 |
| chr4_78093919_78094070 | 0.34 |
CCNG2 |
cyclin G2 |
14415 |
0.22 |
| chr1_117908214_117909192 | 0.34 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
1368 |
0.58 |
| chr11_27448938_27449230 | 0.34 |
RP11-159H22.2 |
|
44192 |
0.13 |
| chr11_33298506_33298657 | 0.34 |
HIPK3 |
homeodomain interacting protein kinase 3 |
18704 |
0.21 |
| chr2_216635749_216635900 | 0.34 |
ENSG00000212055 |
. |
107818 |
0.07 |
| chr7_47515286_47515437 | 0.34 |
TNS3 |
tensin 3 |
5522 |
0.34 |
| chr9_97628790_97628941 | 0.34 |
RP11-49O14.2 |
|
33846 |
0.14 |
| chr18_22907594_22907986 | 0.34 |
ZNF521 |
zinc finger protein 521 |
23367 |
0.23 |
| chr5_39420581_39420732 | 0.33 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
4314 |
0.32 |
| chr16_22409641_22409792 | 0.33 |
CDR2 |
cerebellar degeneration-related protein 2, 62kDa |
23305 |
0.12 |
| chr16_77392856_77393007 | 0.33 |
ADAMTS18 |
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
69675 |
0.1 |
| chr5_173958937_173959224 | 0.33 |
MSX2 |
msh homeobox 2 |
192456 |
0.03 |
| chr2_218354637_218354788 | 0.33 |
ENSG00000251982 |
. |
238433 |
0.02 |
| chr18_8615630_8615858 | 0.33 |
RAB12 |
RAB12, member RAS oncogene family |
6301 |
0.21 |
| chr4_157699893_157700044 | 0.33 |
RP11-154F14.2 |
|
62543 |
0.12 |
| chr14_71368302_71368567 | 0.32 |
PCNX |
pecanex homolog (Drosophila) |
5688 |
0.31 |
| chr10_80865141_80865370 | 0.32 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
36463 |
0.19 |
| chr13_60354193_60354344 | 0.32 |
DIAPH3-AS1 |
DIAPH3 antisense RNA 1 |
232617 |
0.02 |
| chr14_61244389_61244540 | 0.32 |
MNAT1 |
MNAT CDK-activating kinase assembly factor 1 |
30578 |
0.19 |
| chr10_76050762_76050913 | 0.32 |
ADK |
adenosine kinase |
114316 |
0.06 |
| chr7_5458267_5458922 | 0.32 |
AC093620.5 |
|
864 |
0.36 |
| chr8_18815764_18816021 | 0.32 |
ENSG00000201157 |
. |
21237 |
0.2 |
| chr18_18743331_18743728 | 0.32 |
ENSG00000251886 |
. |
43062 |
0.15 |
| chr3_10412398_10412669 | 0.32 |
ENSG00000216135 |
. |
23713 |
0.13 |
| chr1_210551647_210551798 | 0.32 |
ENSG00000200972 |
. |
4111 |
0.28 |
| chr2_123391337_123391637 | 0.32 |
TSN |
translin |
877740 |
0.0 |
| chr13_109567579_109568598 | 0.31 |
MYO16 |
myosin XVI |
29571 |
0.26 |
| chr10_45473918_45474188 | 0.31 |
C10orf10 |
chromosome 10 open reading frame 10 |
184 |
0.91 |
| chr4_41014654_41014947 | 0.31 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
1602 |
0.41 |
| chr3_149119024_149119417 | 0.31 |
TM4SF1-AS1 |
TM4SF1 antisense RNA 1 |
23214 |
0.16 |
| chr1_39575783_39575934 | 0.31 |
MACF1 |
microtubule-actin crosslinking factor 1 |
4775 |
0.2 |
| chr1_7870325_7870476 | 0.31 |
RP3-467L1.4 |
|
17002 |
0.14 |
| chr12_29895057_29895338 | 0.31 |
TMTC1 |
transmembrane and tetratricopeptide repeat containing 1 |
41534 |
0.21 |
| chr10_44958607_44958758 | 0.31 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
78142 |
0.11 |
| chr4_170926232_170926383 | 0.31 |
MFAP3L |
microfibrillar-associated protein 3-like |
1372 |
0.48 |
| chr3_100762310_100762461 | 0.31 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
50026 |
0.16 |
| chr21_35705528_35705679 | 0.31 |
ENSG00000266007 |
. |
28173 |
0.13 |
| chr15_71347018_71347169 | 0.31 |
THSD4 |
thrombospondin, type I, domain containing 4 |
42198 |
0.16 |
| chr2_202663671_202663822 | 0.31 |
CDK15 |
cyclin-dependent kinase 15 |
7352 |
0.16 |
| chr5_37831903_37832054 | 0.30 |
GDNF |
glial cell derived neurotrophic factor |
3032 |
0.32 |
| chr22_36019288_36019458 | 0.30 |
MB |
myoglobin |
28 |
0.98 |
| chr4_47566719_47567004 | 0.30 |
AC092597.3 |
|
4585 |
0.3 |
| chr3_143489302_143489453 | 0.30 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
77923 |
0.12 |
| chr1_86034688_86034839 | 0.30 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
9170 |
0.19 |
| chr1_95258755_95258906 | 0.30 |
SLC44A3 |
solute carrier family 44, member 3 |
27068 |
0.18 |
| chr17_30049392_30049543 | 0.30 |
ENSG00000202026 |
. |
8695 |
0.23 |
| chr8_124683811_124683962 | 0.30 |
KLHL38 |
kelch-like family member 38 |
18696 |
0.19 |
| chr12_46949120_46949271 | 0.30 |
SLC38A2 |
solute carrier family 38, member 2 |
182545 |
0.03 |
| chr12_30947570_30947865 | 0.30 |
CAPRIN2 |
caprin family member 2 |
39832 |
0.19 |
| chr4_150041484_150041635 | 0.30 |
ENSG00000239168 |
. |
128774 |
0.06 |
| chr21_36950874_36951025 | 0.30 |
ENSG00000211590 |
. |
142064 |
0.05 |
| chr5_38783257_38783408 | 0.29 |
RP11-122C5.3 |
|
350 |
0.92 |
| chr7_33793687_33793838 | 0.29 |
RP11-89N17.2 |
|
22086 |
0.22 |
| chr9_72659789_72660086 | 0.29 |
MAMDC2 |
MAM domain containing 2 |
1440 |
0.51 |
| chr11_96042019_96042312 | 0.29 |
ENSG00000266192 |
. |
32437 |
0.17 |
| chr2_36589813_36589964 | 0.29 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
6274 |
0.32 |
| chr1_30556903_30557122 | 0.29 |
ENSG00000222787 |
. |
199263 |
0.03 |
| chr7_114627811_114627962 | 0.29 |
MDFIC |
MyoD family inhibitor domain containing |
53962 |
0.18 |
| chr2_114635767_114636012 | 0.29 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
11648 |
0.21 |
| chr12_47353443_47353741 | 0.29 |
PCED1B |
PC-esterase domain containing 1B |
119794 |
0.06 |
| chr1_59479598_59479749 | 0.29 |
JUN |
jun proto-oncogene |
229888 |
0.02 |
| chr17_80355013_80355164 | 0.29 |
RP13-20L14.4 |
|
4170 |
0.11 |
| chr1_94095744_94095895 | 0.29 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
16165 |
0.21 |
| chr12_42958679_42958830 | 0.29 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
24724 |
0.21 |
| chr17_43407056_43407601 | 0.28 |
ENSG00000199953 |
. |
2465 |
0.22 |
| chr18_74188097_74188669 | 0.28 |
ZNF516 |
zinc finger protein 516 |
14352 |
0.17 |
| chr8_99014594_99014745 | 0.28 |
MATN2 |
matrilin 2 |
14103 |
0.15 |
| chr12_47165529_47165680 | 0.28 |
SLC38A4 |
solute carrier family 38, member 4 |
2517 |
0.44 |
| chr17_26673764_26673915 | 0.28 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
6407 |
0.07 |
| chr8_115953594_115953745 | 0.28 |
TRPS1 |
trichorhinophalangeal syndrome I |
550779 |
0.0 |
| chr5_73497876_73498136 | 0.28 |
ENSG00000222551 |
. |
69584 |
0.13 |
| chr1_161880198_161880487 | 0.28 |
OLFML2B |
olfactomedin-like 2B |
74680 |
0.09 |
| chr8_129179760_129180073 | 0.28 |
ENSG00000221261 |
. |
17554 |
0.23 |
| chr11_35008663_35008941 | 0.28 |
PDHX |
pyruvate dehydrogenase complex, component X |
9471 |
0.23 |
| chr15_99398645_99398796 | 0.28 |
IGF1R |
insulin-like growth factor 1 receptor |
34850 |
0.16 |
| chr3_194880738_194881131 | 0.28 |
XXYLT1-AS2 |
XXYLT1 antisense RNA 2 |
12314 |
0.14 |
| chr4_169555148_169555325 | 0.28 |
PALLD |
palladin, cytoskeletal associated protein |
2468 |
0.31 |
| chr22_32955246_32955397 | 0.28 |
LL22NC03-104C7.1 |
|
23965 |
0.19 |
| chr22_38735164_38735315 | 0.28 |
CSNK1E |
casein kinase 1, epsilon |
21150 |
0.13 |
| chr6_150177550_150177701 | 0.28 |
RP11-350J20.12 |
|
4027 |
0.14 |
| chr13_44819054_44819205 | 0.28 |
SMIM2 |
small integral membrane protein 2 |
83736 |
0.09 |
| chr18_9348065_9348216 | 0.28 |
TWSG1 |
twisted gastrulation BMP signaling modulator 1 |
13292 |
0.2 |
| chr9_22683470_22683621 | 0.28 |
DMRTA1 |
DMRT-like family A1 |
236705 |
0.02 |
| chr10_73627738_73628072 | 0.28 |
PSAP |
prosaposin |
16779 |
0.19 |
| chr12_6198255_6198797 | 0.27 |
ENSG00000240533 |
. |
6610 |
0.23 |
| chr17_4109949_4110100 | 0.27 |
ENSG00000207390 |
. |
13961 |
0.13 |
| chr8_57633478_57633629 | 0.27 |
ENSG00000206975 |
. |
196090 |
0.03 |
| chr3_171998152_171998303 | 0.27 |
AC092964.2 |
Uncharacterized protein |
21459 |
0.19 |
| chr20_52353487_52354058 | 0.27 |
ENSG00000238468 |
. |
68475 |
0.12 |
| chr20_9389511_9389662 | 0.27 |
PLCB4 |
phospholipase C, beta 4 |
101139 |
0.07 |
| chr10_124066964_124067115 | 0.27 |
BTBD16 |
BTB (POZ) domain containing 16 |
36218 |
0.14 |
| chr7_78159625_78159776 | 0.27 |
MAGI2 |
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
240696 |
0.02 |
| chr7_42184494_42184645 | 0.27 |
GLI3 |
GLI family zinc finger 3 |
82751 |
0.12 |
| chr18_9017327_9018234 | 0.27 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
84848 |
0.08 |
| chr7_95239146_95239504 | 0.27 |
AC002451.3 |
|
217 |
0.94 |
| chr3_47435871_47436022 | 0.27 |
PTPN23 |
protein tyrosine phosphatase, non-receptor type 23 |
13436 |
0.15 |
| chr8_104117881_104118032 | 0.27 |
ENSG00000265667 |
. |
13007 |
0.13 |
| chr3_187990037_187990188 | 0.27 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
32460 |
0.22 |
| chr6_12577482_12577633 | 0.27 |
PHACTR1 |
phosphatase and actin regulator 1 |
140336 |
0.05 |
| chr7_42147315_42147466 | 0.27 |
GLI3 |
GLI family zinc finger 3 |
119930 |
0.07 |
| chrX_71489517_71489668 | 0.27 |
RPS4X |
ribosomal protein S4, X-linked |
7506 |
0.14 |
| chr4_55147977_55148291 | 0.27 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
38392 |
0.21 |
| chr5_143075483_143075634 | 0.27 |
ENSG00000266478 |
. |
16133 |
0.26 |
| chr11_66739265_66739416 | 0.27 |
C11orf86 |
chromosome 11 open reading frame 86 |
3408 |
0.16 |
| chr12_13043607_13043874 | 0.27 |
GPRC5A |
G protein-coupled receptor, family C, group 5, member A |
24 |
0.97 |
| chr14_51242480_51242631 | 0.27 |
NIN |
ninein (GSK3B interacting protein) |
9042 |
0.17 |
| chr4_13987753_13987904 | 0.27 |
ENSG00000252092 |
. |
327577 |
0.01 |
| chr12_93483466_93483617 | 0.27 |
ENSG00000221617 |
. |
7486 |
0.23 |
| chr1_61803481_61803729 | 0.27 |
NFIA |
nuclear factor I/A |
14517 |
0.3 |
| chr17_30429974_30430125 | 0.27 |
RP11-640N20.8 |
|
8098 |
0.14 |
| chr4_96471329_96471480 | 0.27 |
UNC5C |
unc-5 homolog C (C. elegans) |
1047 |
0.43 |
| chr5_134475910_134476061 | 0.27 |
PITX1 |
paired-like homeodomain 1 |
105482 |
0.06 |
| chr10_98727488_98727639 | 0.27 |
C10orf12 |
chromosome 10 open reading frame 12 |
13478 |
0.19 |
| chr4_88949160_88949362 | 0.26 |
PKD2 |
polycystic kidney disease 2 (autosomal dominant) |
15152 |
0.19 |
| chr1_183009603_183009956 | 0.26 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
17184 |
0.19 |
| chr4_157242892_157243172 | 0.26 |
ENSG00000221189 |
. |
25234 |
0.27 |
| chr2_47243580_47243731 | 0.26 |
AC093732.1 |
|
24008 |
0.16 |
| chr12_52078091_52078242 | 0.26 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
2041 |
0.42 |
| chr2_218715975_218716307 | 0.26 |
TNS1 |
tensin 1 |
9258 |
0.27 |
| chr2_27516048_27516199 | 0.26 |
TRIM54 |
tripartite motif containing 54 |
10793 |
0.08 |
| chr12_66290144_66290295 | 0.26 |
RP11-366L20.2 |
Uncharacterized protein |
14272 |
0.17 |
| chr4_39616282_39616433 | 0.26 |
ENSG00000252796 |
. |
6275 |
0.19 |
| chr12_67753116_67753506 | 0.26 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
60559 |
0.16 |
| chr11_74724874_74725025 | 0.26 |
NEU3 |
sialidase 3 (membrane sialidase) |
24959 |
0.14 |
| chr3_59752377_59752528 | 0.26 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
205131 |
0.03 |
| chr12_123448783_123448934 | 0.26 |
ABCB9 |
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
2149 |
0.23 |
| chr6_117813949_117814100 | 0.26 |
DCBLD1 |
discoidin, CUB and LCCL domain containing 1 |
10199 |
0.18 |
| chr1_39608036_39608283 | 0.26 |
ENSG00000222378 |
. |
11809 |
0.17 |
| chr18_55317311_55317462 | 0.26 |
RP11-35G9.3 |
|
10699 |
0.16 |
| chr20_1796404_1796650 | 0.26 |
SIRPA |
signal-regulatory protein alpha |
78627 |
0.09 |
| chr6_52409613_52409764 | 0.26 |
TRAM2 |
translocation associated membrane protein 2 |
32025 |
0.2 |
| chr6_116879356_116879507 | 0.26 |
FAM26D |
family with sequence similarity 26, member D |
4518 |
0.16 |
| chr3_37156478_37156973 | 0.25 |
ENSG00000206645 |
. |
22000 |
0.14 |
| chr5_67752834_67752985 | 0.25 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
164513 |
0.04 |
| chr5_119935266_119935417 | 0.25 |
PRR16 |
proline rich 16 |
17442 |
0.29 |
| chr10_3259828_3259979 | 0.25 |
PITRM1 |
pitrilysin metallopeptidase 1 |
44900 |
0.18 |
| chr12_22886601_22886752 | 0.25 |
ETNK1 |
ethanolamine kinase 1 |
108385 |
0.07 |
| chr10_6764254_6764548 | 0.25 |
PRKCQ |
protein kinase C, theta |
142138 |
0.05 |
| chr10_112083656_112083807 | 0.25 |
SMNDC1 |
survival motor neuron domain containing 1 |
19022 |
0.23 |
| chr2_10113482_10113633 | 0.25 |
GRHL1 |
grainyhead-like 1 (Drosophila) |
21730 |
0.16 |
| chr4_38084935_38085086 | 0.25 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
37518 |
0.21 |
| chr10_71206366_71206517 | 0.25 |
TSPAN15 |
tetraspanin 15 |
4788 |
0.23 |
| chr1_45412695_45413130 | 0.25 |
ENSG00000253039 |
. |
14917 |
0.13 |
| chr9_115947226_115947377 | 0.25 |
SLC31A2 |
solute carrier family 31 (copper transporter), member 2 |
34079 |
0.13 |
| chr9_91610255_91610406 | 0.25 |
S1PR3 |
sphingosine-1-phosphate receptor 3 |
1091 |
0.53 |
| chr1_170001529_170001680 | 0.25 |
KIFAP3 |
kinesin-associated protein 3 |
6562 |
0.21 |
| chr3_149738282_149738433 | 0.25 |
PFN2 |
profilin 2 |
2241 |
0.29 |
| chr10_31934113_31934264 | 0.25 |
ENSG00000222412 |
. |
110876 |
0.07 |
| chr5_60625410_60625800 | 0.25 |
ZSWIM6 |
zinc finger, SWIM-type containing 6 |
2495 |
0.42 |
| chr6_16676788_16676939 | 0.25 |
RP1-151F17.1 |
|
84506 |
0.1 |
| chr9_90215657_90216062 | 0.24 |
DAPK1-IT1 |
DAPK1 intronic transcript 1 (non-protein coding) |
47490 |
0.16 |
| chr6_140087291_140087442 | 0.24 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
391609 |
0.01 |
| chr7_142171579_142172312 | 0.24 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
182336 |
0.03 |
| chr2_12959099_12960371 | 0.24 |
ENSG00000264370 |
. |
82242 |
0.11 |
| chr6_15328781_15329139 | 0.24 |
ENSG00000201519 |
. |
4257 |
0.25 |
| chr3_31419139_31419290 | 0.24 |
ENSG00000238727 |
. |
18215 |
0.28 |
| chr9_6433707_6433858 | 0.24 |
UHRF2 |
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
12751 |
0.18 |
| chr9_113597331_113597482 | 0.24 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
59779 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.3 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.1 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.2 | GO:0035929 | steroid hormone secretion(GO:0035929) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.1 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.2 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0061047 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.0 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0061042 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0010662 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.3 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.1 | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis(GO:0060502) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.0 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0014848 | urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.1 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.0 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.0 | 0.0 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.0 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.2 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:1903020 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.0 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.2 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.2 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.2 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.0 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.0 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |