Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
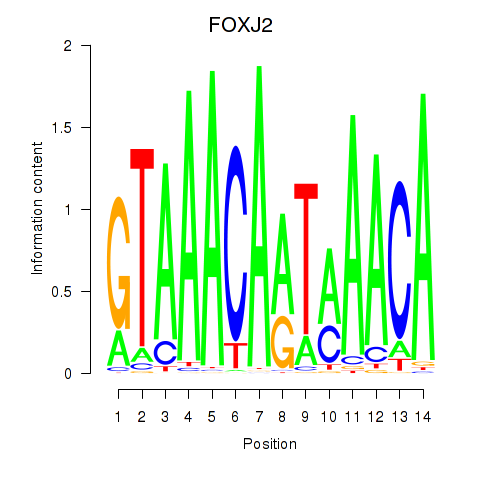
Results for FOXJ2
Z-value: 0.90
Transcription factors associated with FOXJ2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXJ2
|
ENSG00000065970.4 | forkhead box J2 |
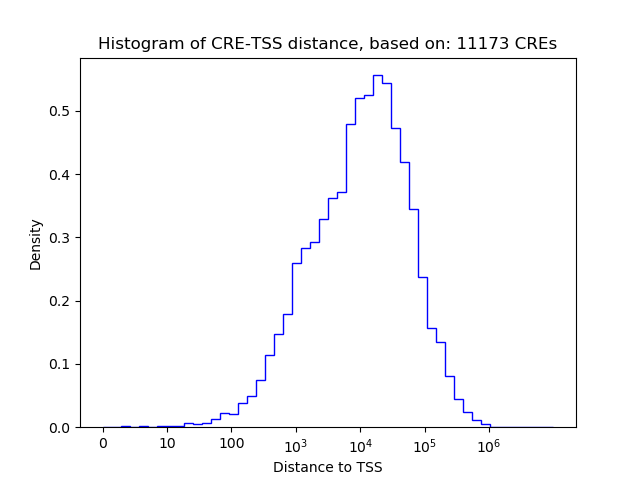
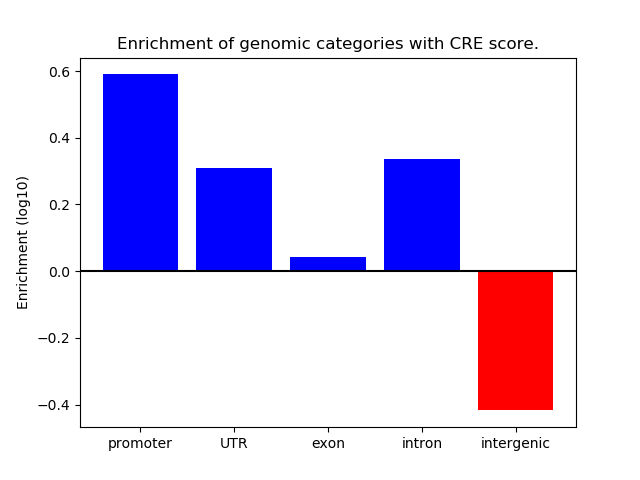
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_8184554_8184838 | FOXJ2 | 603 | 0.697667 | -0.60 | 8.7e-02 | Click! |
| chr12_8171134_8171285 | FOXJ2 | 14090 | 0.151001 | 0.60 | 9.0e-02 | Click! |
| chr12_8188502_8188653 | FOXJ2 | 2621 | 0.229622 | 0.55 | 1.3e-01 | Click! |
| chr12_8190705_8190938 | FOXJ2 | 4865 | 0.174773 | 0.54 | 1.3e-01 | Click! |
| chr12_8182937_8183088 | FOXJ2 | 2287 | 0.254019 | 0.48 | 1.9e-01 | Click! |
Activity of the FOXJ2 motif across conditions
Conditions sorted by the z-value of the FOXJ2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
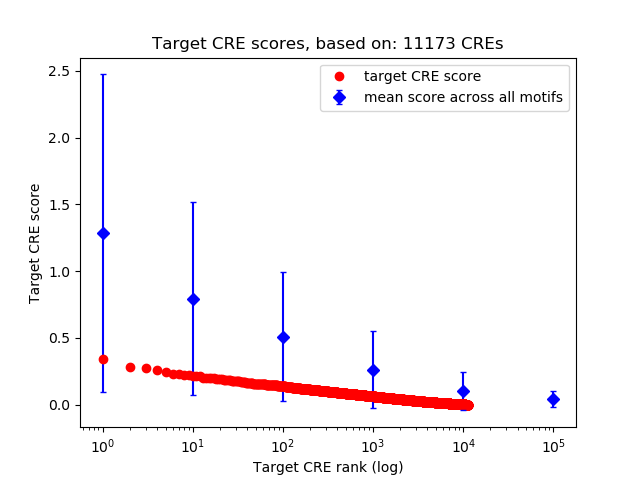
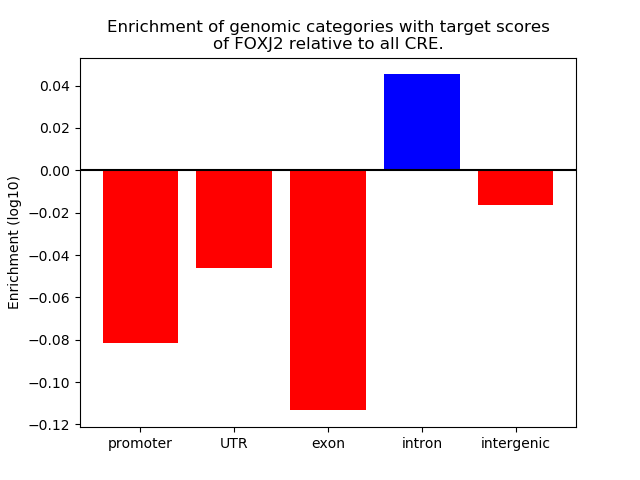
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_26704220_26704518 | 0.35 |
C7orf71 |
chromosome 7 open reading frame 71 |
26879 |
0.23 |
| chr3_187627904_187628178 | 0.28 |
BCL6 |
B-cell CLL/lymphoma 6 |
164526 |
0.04 |
| chr11_64078371_64078522 | 0.28 |
ESRRA |
estrogen-related receptor alpha |
3310 |
0.08 |
| chr12_57077760_57078152 | 0.26 |
PTGES3 |
prostaglandin E synthase 3 (cytosolic) |
3990 |
0.14 |
| chr12_7123219_7123370 | 0.24 |
LPCAT3 |
lysophosphatidylcholine acyltransferase 3 |
2520 |
0.14 |
| chr3_31975446_31975713 | 0.23 |
OSBPL10 |
oxysterol binding protein-like 10 |
47213 |
0.14 |
| chr18_72895850_72896001 | 0.23 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
21543 |
0.23 |
| chr11_32916634_32916991 | 0.22 |
QSER1 |
glutamine and serine rich 1 |
2088 |
0.38 |
| chr7_26900574_26900830 | 0.22 |
SKAP2 |
src kinase associated phosphoprotein 2 |
3463 |
0.33 |
| chr10_116153047_116153198 | 0.22 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
11117 |
0.23 |
| chr2_110402358_110402509 | 0.21 |
SOWAHC |
sosondowah ankyrin repeat domain family member C |
30522 |
0.16 |
| chr8_122360906_122361134 | 0.21 |
ENSG00000221644 |
. |
161888 |
0.04 |
| chr3_172281504_172281789 | 0.20 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
40349 |
0.16 |
| chr18_74197341_74197735 | 0.20 |
ZNF516 |
zinc finger protein 516 |
5197 |
0.19 |
| chr1_24284423_24284685 | 0.20 |
CNR2 |
cannabinoid receptor 2 (macrophage) |
995 |
0.32 |
| chr6_116420339_116420630 | 0.20 |
NT5DC1 |
5'-nucleotidase domain containing 1 |
1528 |
0.38 |
| chr6_138913719_138913963 | 0.20 |
NHSL1 |
NHS-like 1 |
20164 |
0.24 |
| chr22_38294132_38294330 | 0.20 |
ENSG00000207227 |
. |
6877 |
0.1 |
| chr1_43411413_43411601 | 0.19 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
12993 |
0.18 |
| chr20_1784583_1784806 | 0.19 |
SIRPA |
signal-regulatory protein alpha |
90460 |
0.07 |
| chr18_46925702_46925885 | 0.19 |
DYM |
dymeclin |
20774 |
0.15 |
| chr3_112365839_112365990 | 0.19 |
CCDC80 |
coiled-coil domain containing 80 |
5767 |
0.27 |
| chr14_91607015_91607166 | 0.19 |
C14orf159 |
chromosome 14 open reading frame 159 |
7032 |
0.16 |
| chr6_37591161_37591419 | 0.19 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
22860 |
0.22 |
| chr8_66629977_66630128 | 0.19 |
RP11-707M3.3 |
|
3039 |
0.28 |
| chr3_51534436_51534590 | 0.18 |
VPRBP |
Vpr (HIV-1) binding protein |
503 |
0.82 |
| chr4_143345126_143345417 | 0.18 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
7141 |
0.35 |
| chr3_12435782_12435933 | 0.18 |
PPARG |
peroxisome proliferator-activated receptor gamma |
42886 |
0.15 |
| chr20_17836751_17837016 | 0.18 |
ENSG00000221220 |
. |
13656 |
0.2 |
| chr12_14667928_14668079 | 0.18 |
RP11-502N13.2 |
|
15082 |
0.16 |
| chr11_92585503_92585654 | 0.18 |
FAT3 |
FAT atypical cadherin 3 |
8072 |
0.28 |
| chr14_66239499_66239650 | 0.18 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
103441 |
0.08 |
| chr14_68714875_68715122 | 0.17 |
ENSG00000243546 |
. |
11742 |
0.27 |
| chr3_23366935_23367198 | 0.17 |
ENSG00000206728 |
. |
51768 |
0.15 |
| chr19_17523343_17523496 | 0.17 |
MVB12A |
multivesicular body subunit 12A |
6465 |
0.08 |
| chr6_16652590_16652871 | 0.17 |
RP1-151F17.1 |
|
108639 |
0.07 |
| chr12_111807867_111808018 | 0.17 |
FAM109A |
family with sequence similarity 109, member A |
1017 |
0.54 |
| chr11_46861994_46862145 | 0.17 |
CKAP5 |
cytoskeleton associated protein 5 |
5711 |
0.17 |
| chr19_47371603_47371754 | 0.17 |
AP2S1 |
adaptor-related protein complex 2, sigma 1 subunit |
17429 |
0.16 |
| chr4_105416352_105416699 | 0.17 |
CXXC4 |
CXXC finger protein 4 |
474 |
0.81 |
| chr11_10609627_10609795 | 0.16 |
MRVI1-AS1 |
MRVI1 antisense RNA 1 |
5033 |
0.19 |
| chr22_18539461_18539695 | 0.16 |
XXbac-B476C20.9 |
|
20985 |
0.13 |
| chr5_39148697_39148939 | 0.16 |
AC008964.1 |
|
43460 |
0.16 |
| chr22_43322871_43323022 | 0.16 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
18933 |
0.21 |
| chr3_185888596_185888848 | 0.16 |
ETV5 |
ets variant 5 |
60615 |
0.13 |
| chr6_150463083_150463314 | 0.16 |
PPP1R14C |
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
1014 |
0.56 |
| chr4_90756774_90756958 | 0.16 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
20 |
0.94 |
| chr9_124116966_124117153 | 0.16 |
AL161784.1 |
Uncharacterized protein |
13434 |
0.14 |
| chr5_127295498_127295826 | 0.16 |
SLC12A2 |
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
123796 |
0.06 |
| chr2_29339694_29340047 | 0.16 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
1191 |
0.44 |
| chr2_128554283_128554484 | 0.16 |
ENSG00000201470 |
. |
2189 |
0.25 |
| chr17_6581080_6581264 | 0.16 |
SLC13A5 |
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
13031 |
0.1 |
| chr10_60935903_60936054 | 0.16 |
PHYHIPL |
phytanoyl-CoA 2-hydroxylase interacting protein-like |
372 |
0.92 |
| chr10_74989869_74990020 | 0.16 |
DNAJC9-AS1 |
DNAJC9 antisense RNA 1 |
17174 |
0.09 |
| chr1_32040359_32040510 | 0.16 |
RP11-73M7.1 |
|
740 |
0.54 |
| chr6_116772996_116773147 | 0.16 |
ENSG00000265516 |
. |
5914 |
0.14 |
| chr15_59461940_59462091 | 0.16 |
ENSG00000253030 |
. |
1446 |
0.28 |
| chrX_129254186_129254397 | 0.15 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
9600 |
0.2 |
| chr10_14823935_14824305 | 0.15 |
FAM107B |
family with sequence similarity 107, member B |
7224 |
0.22 |
| chr6_130924438_130924589 | 0.15 |
ENSG00000202438 |
. |
29256 |
0.24 |
| chr7_41548338_41548489 | 0.15 |
INHBA-AS1 |
INHBA antisense RNA 1 |
185101 |
0.03 |
| chr14_71349673_71349956 | 0.15 |
PCNX |
pecanex homolog (Drosophila) |
24308 |
0.24 |
| chr14_103838670_103838952 | 0.15 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
12918 |
0.15 |
| chr12_680792_680943 | 0.15 |
RP5-1154L15.2 |
|
14785 |
0.13 |
| chr10_97047146_97047394 | 0.15 |
PDLIM1 |
PDZ and LIM domain 1 |
3511 |
0.3 |
| chr4_38387684_38387835 | 0.15 |
ENSG00000221495 |
. |
147525 |
0.04 |
| chr7_20830813_20830964 | 0.15 |
SP8 |
Sp8 transcription factor |
4383 |
0.36 |
| chr10_102671101_102671252 | 0.15 |
FAM178A |
family with sequence similarity 178, member A |
1150 |
0.31 |
| chr1_81697056_81697207 | 0.15 |
ENSG00000223026 |
. |
20343 |
0.27 |
| chr16_57414802_57415099 | 0.15 |
CX3CL1 |
chemokine (C-X3-C motif) ligand 1 |
4066 |
0.16 |
| chr8_18672395_18672546 | 0.15 |
ENSG00000251963 |
. |
5334 |
0.24 |
| chr9_124375460_124375641 | 0.15 |
RP11-524G24.2 |
|
37057 |
0.16 |
| chr4_159705947_159706098 | 0.15 |
ENSG00000206703 |
. |
9826 |
0.19 |
| chr1_214666364_214666654 | 0.15 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
28363 |
0.24 |
| chr4_15033017_15033168 | 0.15 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
22665 |
0.25 |
| chr3_70890218_70890369 | 0.15 |
ENSG00000206939 |
. |
32792 |
0.2 |
| chr15_99211968_99212377 | 0.15 |
IGF1R |
insulin-like growth factor 1 receptor |
19899 |
0.21 |
| chr9_91047692_91047843 | 0.15 |
SPIN1 |
spindlin 1 |
44421 |
0.17 |
| chr5_170738876_170739099 | 0.15 |
TLX3 |
T-cell leukemia homeobox 3 |
2699 |
0.26 |
| chr12_72240351_72240502 | 0.15 |
TBC1D15 |
TBC1 domain family, member 15 |
6868 |
0.27 |
| chr13_110651731_110651882 | 0.15 |
ENSG00000201161 |
. |
84363 |
0.1 |
| chr7_156945282_156945433 | 0.15 |
UBE3C |
ubiquitin protein ligase E3C |
13468 |
0.22 |
| chr4_85602887_85603189 | 0.15 |
ENSG00000239466 |
. |
6133 |
0.27 |
| chr19_23437287_23437525 | 0.15 |
ZNF724P |
zinc finger protein 724, pseudogene |
4214 |
0.29 |
| chr4_143172838_143172989 | 0.14 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
54072 |
0.19 |
| chr19_18490843_18491034 | 0.14 |
GDF15 |
growth differentiation factor 15 |
2042 |
0.16 |
| chr9_80539292_80539443 | 0.14 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
101452 |
0.08 |
| chr8_8820590_8820741 | 0.14 |
ENSG00000207244 |
. |
1580 |
0.39 |
| chr3_43334090_43334357 | 0.14 |
SNRK |
SNF related kinase |
6145 |
0.2 |
| chr15_68376694_68376845 | 0.14 |
PIAS1 |
protein inhibitor of activated STAT, 1 |
29855 |
0.22 |
| chr18_9596078_9596229 | 0.14 |
RP11-881L2.1 |
|
8769 |
0.16 |
| chrX_153265369_153265652 | 0.14 |
IRAK1 |
interleukin-1 receptor-associated kinase 1 |
14187 |
0.09 |
| chr20_50108720_50109319 | 0.14 |
ENSG00000266761 |
. |
39505 |
0.18 |
| chr17_4262517_4262743 | 0.14 |
ENSG00000263882 |
. |
5118 |
0.18 |
| chr6_15381784_15381935 | 0.14 |
JARID2 |
jumonji, AT rich interactive domain 2 |
19230 |
0.22 |
| chr8_102038362_102038931 | 0.14 |
ENSG00000252736 |
. |
63375 |
0.1 |
| chr16_69431934_69432085 | 0.14 |
TERF2 |
telomeric repeat binding factor 2 |
10465 |
0.13 |
| chr3_42121230_42121496 | 0.14 |
TRAK1 |
trafficking protein, kinesin binding 1 |
11199 |
0.27 |
| chr10_114854381_114854621 | 0.14 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
49178 |
0.18 |
| chr5_64505728_64505879 | 0.14 |
ENSG00000207439 |
. |
86607 |
0.1 |
| chr2_112789798_112789949 | 0.14 |
MERTK |
c-mer proto-oncogene tyrosine kinase |
12883 |
0.21 |
| chr9_4661316_4661551 | 0.14 |
PPAPDC2 |
phosphatidic acid phosphatase type 2 domain containing 2 |
865 |
0.57 |
| chr17_60868423_60868627 | 0.14 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
15476 |
0.23 |
| chr15_68177525_68177676 | 0.14 |
ENSG00000206625 |
. |
45217 |
0.17 |
| chr15_55881471_55881622 | 0.14 |
PYGO1 |
pygopus family PHD finger 1 |
401 |
0.89 |
| chr2_43242041_43242192 | 0.14 |
ENSG00000207087 |
. |
76516 |
0.11 |
| chr8_18066990_18067251 | 0.14 |
NAT1 |
N-acetyltransferase 1 (arylamine N-acetyltransferase) |
482 |
0.88 |
| chr9_102682165_102682316 | 0.14 |
RP11-60I3.4 |
|
9938 |
0.18 |
| chr17_39068896_39069135 | 0.14 |
AC004231.2 |
|
8667 |
0.1 |
| chr15_71583507_71583658 | 0.14 |
RP11-592N21.2 |
|
51081 |
0.16 |
| chr20_52353487_52354058 | 0.13 |
ENSG00000238468 |
. |
68475 |
0.12 |
| chr3_18468302_18468610 | 0.13 |
SATB1 |
SATB homeobox 1 |
1595 |
0.42 |
| chr12_22486049_22486200 | 0.13 |
ST8SIA1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
973 |
0.7 |
| chr7_17976053_17976204 | 0.13 |
SNX13 |
sorting nexin 13 |
3963 |
0.31 |
| chr1_70469306_70469482 | 0.13 |
RP11-181B18.1 |
|
18687 |
0.26 |
| chr1_107598936_107599204 | 0.13 |
PRMT6 |
protein arginine methyltransferase 6 |
197 |
0.97 |
| chr1_23668057_23668694 | 0.13 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
2380 |
0.23 |
| chr5_55881573_55881982 | 0.13 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
20282 |
0.21 |
| chr5_169577969_169578120 | 0.13 |
FOXI1 |
forkhead box I1 |
45127 |
0.16 |
| chr13_31774982_31775836 | 0.13 |
B3GALTL |
beta 1,3-galactosyltransferase-like |
1336 |
0.55 |
| chr10_82773983_82774134 | 0.13 |
ENSG00000265990 |
. |
130350 |
0.06 |
| chr1_53739428_53739681 | 0.13 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
1781 |
0.2 |
| chr1_161122819_161122970 | 0.13 |
UFC1 |
ubiquitin-fold modifier conjugating enzyme 1 |
648 |
0.42 |
| chr6_90868910_90869061 | 0.13 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
137476 |
0.04 |
| chr7_5704858_5705042 | 0.13 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
15142 |
0.19 |
| chr19_44087068_44087219 | 0.13 |
PINLYP |
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
1954 |
0.14 |
| chr1_51889386_51889537 | 0.13 |
EPS15 |
epidermal growth factor receptor pathway substrate 15 |
471 |
0.8 |
| chr7_121170101_121170252 | 0.13 |
ENSG00000221690 |
. |
44632 |
0.19 |
| chr6_159571881_159572032 | 0.13 |
RP11-13P5.2 |
|
16429 |
0.19 |
| chr18_7084430_7084600 | 0.13 |
RP11-76K13.3 |
|
7699 |
0.23 |
| chr19_18575570_18575881 | 0.13 |
ELL |
elongation factor RNA polymerase II |
11018 |
0.1 |
| chr9_124118304_124118517 | 0.13 |
STOM |
stomatin |
14073 |
0.14 |
| chr2_219129032_219129405 | 0.13 |
AAMP |
angio-associated, migratory cell protein |
1586 |
0.21 |
| chr17_61501531_61501682 | 0.13 |
RP11-269G24.3 |
|
6844 |
0.14 |
| chr1_218633775_218633926 | 0.13 |
C1orf143 |
chromosome 1 open reading frame 143 |
49588 |
0.16 |
| chr1_60391839_60392206 | 0.13 |
CYP2J2 |
cytochrome P450, family 2, subfamily J, polypeptide 2 |
440 |
0.87 |
| chr9_20320271_20320422 | 0.13 |
ENSG00000221744 |
. |
25344 |
0.21 |
| chr6_7050238_7050389 | 0.13 |
ENSG00000251762 |
. |
9534 |
0.25 |
| chr12_54021012_54021163 | 0.13 |
ATF7 |
activating transcription factor 7 |
888 |
0.44 |
| chr11_43757721_43758198 | 0.13 |
HSD17B12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
55628 |
0.13 |
| chrX_48859192_48859344 | 0.13 |
GRIPAP1 |
GRIP1 associated protein 1 |
593 |
0.56 |
| chr1_41178229_41178380 | 0.13 |
NFYC |
nuclear transcription factor Y, gamma |
3089 |
0.2 |
| chr11_35251646_35251916 | 0.12 |
CD44 |
CD44 molecule (Indian blood group) |
10890 |
0.15 |
| chr1_172328500_172328837 | 0.12 |
ENSG00000206684 |
. |
7012 |
0.18 |
| chr15_90925903_90926054 | 0.12 |
IQGAP1 |
IQ motif containing GTPase activating protein 1 |
5472 |
0.15 |
| chr2_2650026_2650177 | 0.12 |
MYT1L |
myelin transcription factor 1-like |
315135 |
0.01 |
| chr20_19797225_19797376 | 0.12 |
AL121761.2 |
Uncharacterized protein |
58621 |
0.12 |
| chr2_48551769_48552030 | 0.12 |
FOXN2 |
forkhead box N2 |
10050 |
0.22 |
| chr12_69894065_69894216 | 0.12 |
FRS2 |
fibroblast growth factor receptor substrate 2 |
29926 |
0.17 |
| chr22_43559847_43560025 | 0.12 |
TTLL12 |
tubulin tyrosine ligase-like family, member 12 |
7820 |
0.14 |
| chr1_83043773_83044235 | 0.12 |
LPHN2 |
latrophilin 2 |
598431 |
0.0 |
| chr9_111874941_111875092 | 0.12 |
TMEM245 |
transmembrane protein 245 |
7209 |
0.2 |
| chr1_172795013_172795164 | 0.12 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
166930 |
0.03 |
| chr2_61193206_61193983 | 0.12 |
ENSG00000222251 |
. |
32293 |
0.14 |
| chr15_83513364_83513515 | 0.12 |
HOMER2 |
homer homolog 2 (Drosophila) |
5040 |
0.17 |
| chr17_74259263_74259519 | 0.12 |
UBALD2 |
UBA-like domain containing 2 |
1892 |
0.24 |
| chr2_25584992_25585274 | 0.12 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
19674 |
0.2 |
| chr6_139090481_139090632 | 0.12 |
CCDC28A |
coiled-coil domain containing 28A |
4101 |
0.24 |
| chr6_16737522_16738057 | 0.12 |
RP1-151F17.1 |
|
23580 |
0.22 |
| chr20_56827001_56827171 | 0.12 |
PPP4R1L |
protein phosphatase 4, regulatory subunit 1-like |
264 |
0.93 |
| chr5_158632936_158633295 | 0.12 |
RNF145 |
ring finger protein 145 |
1527 |
0.36 |
| chr5_68798177_68798328 | 0.12 |
RP11-241G9.3 |
|
41 |
0.96 |
| chr11_46130483_46130634 | 0.12 |
ENSG00000263539 |
. |
4211 |
0.24 |
| chr6_170596098_170596249 | 0.12 |
DLL1 |
delta-like 1 (Drosophila) |
3388 |
0.21 |
| chr7_32531022_32531199 | 0.12 |
LSM5 |
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
1080 |
0.58 |
| chr15_59642926_59643113 | 0.12 |
RP11-356M20.3 |
|
9069 |
0.13 |
| chr6_2546230_2546381 | 0.12 |
C6orf195 |
chromosome 6 open reading frame 195 |
88532 |
0.09 |
| chr10_116616195_116616346 | 0.12 |
FAM160B1 |
family with sequence similarity 160, member B1 |
4280 |
0.32 |
| chr2_191583791_191583942 | 0.12 |
AC006460.2 |
|
10424 |
0.24 |
| chr5_17235667_17236039 | 0.12 |
ENSG00000252908 |
. |
4867 |
0.21 |
| chr12_10794215_10794366 | 0.12 |
STYK1 |
serine/threonine/tyrosine kinase 1 |
12053 |
0.16 |
| chr10_35231257_35231408 | 0.12 |
ENSG00000222909 |
. |
565 |
0.79 |
| chr9_132733431_132733683 | 0.12 |
FNBP1 |
formin binding protein 1 |
23589 |
0.17 |
| chr4_88025923_88026419 | 0.12 |
AFF1 |
AF4/FMR2 family, member 1 |
55367 |
0.15 |
| chr5_88047186_88047337 | 0.12 |
CTC-467M3.2 |
|
58793 |
0.12 |
| chr15_71118836_71119061 | 0.12 |
RP11-138H8.6 |
|
23161 |
0.14 |
| chr9_124706762_124707039 | 0.12 |
RP11-244O19.1 |
|
125169 |
0.05 |
| chr9_128649834_128649985 | 0.12 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
22359 |
0.27 |
| chr1_186342452_186343004 | 0.12 |
TPR |
translocated promoter region, nuclear basket protein |
1729 |
0.26 |
| chr1_173145333_173145484 | 0.12 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
29289 |
0.23 |
| chr8_28570826_28570977 | 0.12 |
EXTL3 |
exostosin-like glycosyltransferase 3 |
3026 |
0.25 |
| chr1_203653860_203654011 | 0.12 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
1998 |
0.36 |
| chr2_208147717_208147868 | 0.12 |
ENSG00000221628 |
. |
13644 |
0.22 |
| chr7_70144187_70144338 | 0.12 |
AUTS2 |
autism susceptibility candidate 2 |
49863 |
0.2 |
| chr3_171072744_171072895 | 0.12 |
ENSG00000222506 |
. |
84496 |
0.09 |
| chr6_116685228_116685657 | 0.12 |
DSE |
dermatan sulfate epimerase |
5568 |
0.22 |
| chr9_37859453_37859604 | 0.12 |
SLC25A51 |
solute carrier family 25, member 51 |
29063 |
0.17 |
| chr15_43781223_43781374 | 0.12 |
TP53BP1 |
tumor protein p53 binding protein 1 |
3986 |
0.18 |
| chr22_41247543_41247694 | 0.12 |
ST13 |
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
2984 |
0.17 |
| chr12_46949120_46949271 | 0.12 |
SLC38A2 |
solute carrier family 38, member 2 |
182545 |
0.03 |
| chr7_25009452_25009603 | 0.12 |
OSBPL3 |
oxysterol binding protein-like 3 |
10233 |
0.27 |
| chr3_141243275_141243445 | 0.12 |
RASA2-IT1 |
RASA2 intronic transcript 1 (non-protein coding) |
615 |
0.79 |
| chr11_32925473_32925624 | 0.12 |
QSER1 |
glutamine and serine rich 1 |
10824 |
0.23 |
| chr18_21406136_21407050 | 0.12 |
LAMA3 |
laminin, alpha 3 |
7324 |
0.26 |
| chr13_40260042_40260269 | 0.12 |
ENSG00000264171 |
. |
21883 |
0.17 |
| chr2_192138781_192138977 | 0.12 |
MYO1B |
myosin IB |
2732 |
0.37 |
| chr14_89297995_89298146 | 0.11 |
TTC8 |
tetratricopeptide repeat domain 8 |
6987 |
0.18 |
| chr3_130681680_130681831 | 0.11 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
4477 |
0.2 |
| chr7_17530312_17530463 | 0.11 |
ENSG00000199473 |
. |
117437 |
0.06 |
| chr5_88340644_88340795 | 0.11 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
79027 |
0.12 |
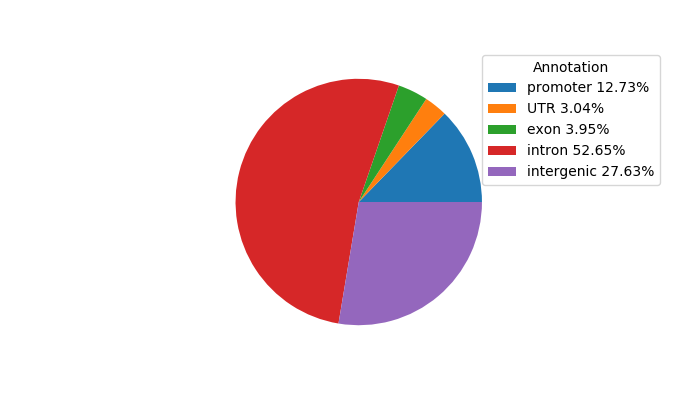
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0033131 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.0 | GO:0048263 | determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0046040 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.0 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0010523 | negative regulation of calcium ion transport into cytosol(GO:0010523) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.0 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |