Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for FOXO3_FOXD2
Z-value: 0.73
Transcription factors associated with FOXO3_FOXD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
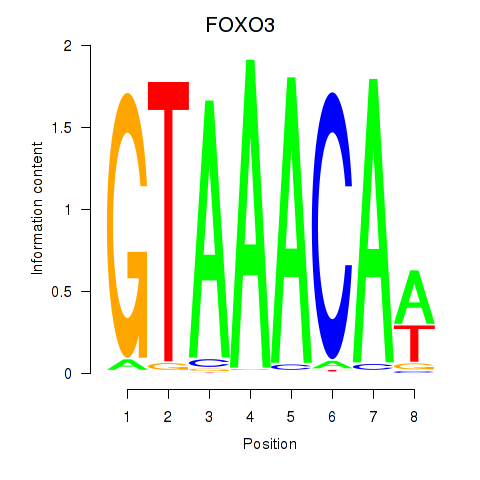
FOXO3
|
ENSG00000118689.10 | forkhead box O3 |
|
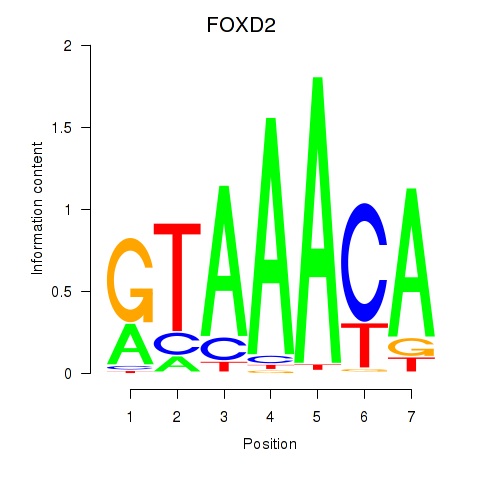
FOXD2
|
ENSG00000186564.5 | forkhead box D2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_47905796_47905947 | FOXD2 | 4182 | 0.177170 | -0.78 | 1.3e-02 | Click! |
| chr1_47903924_47904075 | FOXD2 | 2310 | 0.232951 | 0.62 | 7.6e-02 | Click! |
| chr1_47902342_47903033 | FOXD2 | 998 | 0.452227 | 0.57 | 1.1e-01 | Click! |
| chr1_47906249_47906400 | FOXD2 | 4635 | 0.172242 | -0.56 | 1.2e-01 | Click! |
| chr1_47909973_47910124 | FOXD2 | 8359 | 0.156738 | -0.53 | 1.5e-01 | Click! |
| chr6_108994454_108994605 | FOXO3 | 16980 | 0.275450 | 0.89 | 1.5e-03 | Click! |
| chr6_108885127_108885278 | FOXO3 | 3133 | 0.374122 | -0.88 | 1.7e-03 | Click! |
| chr6_108949983_108950134 | FOXO3 | 27491 | 0.245271 | -0.83 | 5.3e-03 | Click! |
| chr6_108948593_108948758 | FOXO3 | 28874 | 0.240811 | -0.81 | 7.6e-03 | Click! |
| chr6_108917705_108917856 | FOXO3 | 35711 | 0.217656 | 0.76 | 1.7e-02 | Click! |
Activity of the FOXO3_FOXD2 motif across conditions
Conditions sorted by the z-value of the FOXO3_FOXD2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
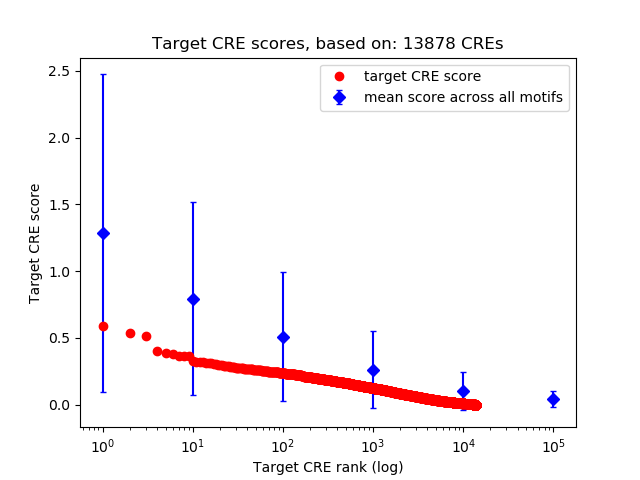
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_9389371_9389522 | 0.59 |
TBL1X |
transducin (beta)-like 1X-linked |
41889 |
0.22 |
| chr22_24879125_24879276 | 0.53 |
ADORA2A-AS1 |
ADORA2A antisense RNA 1 |
11563 |
0.14 |
| chr3_191073989_191074140 | 0.51 |
UTS2B |
urotensin 2B |
25739 |
0.19 |
| chr14_91488374_91488965 | 0.40 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
37809 |
0.15 |
| chr3_133967811_133967962 | 0.39 |
RYK |
receptor-like tyrosine kinase |
1551 |
0.52 |
| chr11_10877596_10878299 | 0.38 |
ZBED5 |
zinc finger, BED-type containing 5 |
1237 |
0.37 |
| chr19_23437287_23437525 | 0.37 |
ZNF724P |
zinc finger protein 724, pseudogene |
4214 |
0.29 |
| chr13_33803306_33803457 | 0.36 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
23238 |
0.2 |
| chr14_61244389_61244540 | 0.36 |
MNAT1 |
MNAT CDK-activating kinase assembly factor 1 |
30578 |
0.19 |
| chr19_34724106_34724375 | 0.33 |
KIAA0355 |
KIAA0355 |
21202 |
0.21 |
| chr1_152415265_152415416 | 0.32 |
CRNN |
cornulin |
28601 |
0.13 |
| chr5_150153974_150154288 | 0.32 |
SMIM3 |
small integral membrane protein 3 |
3377 |
0.19 |
| chr1_185066313_185066464 | 0.32 |
TRMT1L |
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
42304 |
0.13 |
| chr2_70008722_70008880 | 0.31 |
ANXA4 |
annexin A4 |
39565 |
0.14 |
| chr21_39860193_39860344 | 0.31 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
10077 |
0.31 |
| chr4_5035629_5036207 | 0.31 |
CYTL1 |
cytokine-like 1 |
14719 |
0.22 |
| chr5_52627609_52627883 | 0.31 |
FST |
follistatin |
148493 |
0.04 |
| chr4_1759756_1760086 | 0.31 |
TACC3 |
transforming, acidic coiled-coil containing protein 3 |
29790 |
0.12 |
| chr10_99599816_99599967 | 0.30 |
GOLGA7B |
golgin A7 family, member B |
10105 |
0.18 |
| chr9_109203596_109203848 | 0.30 |
ENSG00000200131 |
. |
238536 |
0.02 |
| chr14_53821474_53821625 | 0.29 |
RP11-547D23.1 |
|
201477 |
0.03 |
| chr6_154614018_154614169 | 0.29 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
37122 |
0.22 |
| chr5_37739048_37739199 | 0.29 |
ENSG00000206743 |
. |
38584 |
0.18 |
| chr5_173958937_173959224 | 0.29 |
MSX2 |
msh homeobox 2 |
192456 |
0.03 |
| chr6_64516560_64517136 | 0.29 |
RP11-59D5__B.2 |
|
119 |
0.98 |
| chr7_100781001_100781597 | 0.29 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
10920 |
0.1 |
| chr2_28210866_28211683 | 0.29 |
ENSG00000265321 |
. |
7960 |
0.26 |
| chr3_55525759_55525910 | 0.28 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
1861 |
0.41 |
| chr6_135989503_135989654 | 0.28 |
AHI1 |
Abelson helper integration site 1 |
170664 |
0.03 |
| chr8_94342739_94342890 | 0.28 |
LINC00535 |
long intergenic non-protein coding RNA 535 |
37900 |
0.19 |
| chr13_40260042_40260269 | 0.27 |
ENSG00000264171 |
. |
21883 |
0.17 |
| chr8_66629977_66630128 | 0.27 |
RP11-707M3.3 |
|
3039 |
0.28 |
| chr4_40157960_40158111 | 0.27 |
RP11-395I6.3 |
|
10260 |
0.19 |
| chr11_12147194_12147407 | 0.27 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
4278 |
0.32 |
| chr11_116741651_116741802 | 0.27 |
SIK3-IT1 |
SIK3 intronic transcript 1 (non-protein coding) |
19711 |
0.11 |
| chr12_66271805_66272278 | 0.27 |
RP11-366L20.2 |
Uncharacterized protein |
3317 |
0.24 |
| chr12_10794215_10794366 | 0.27 |
STYK1 |
serine/threonine/tyrosine kinase 1 |
12053 |
0.16 |
| chr1_8728345_8728654 | 0.27 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
12069 |
0.26 |
| chr3_182980866_182981048 | 0.27 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
2133 |
0.32 |
| chr4_74702622_74702974 | 0.27 |
CXCL6 |
chemokine (C-X-C motif) ligand 6 |
441 |
0.79 |
| chr6_47236614_47236765 | 0.27 |
TNFRSF21 |
tumor necrosis factor receptor superfamily, member 21 |
40952 |
0.2 |
| chr3_45576267_45576542 | 0.27 |
LARS2-AS1 |
LARS2 antisense RNA 1 |
25367 |
0.18 |
| chr20_22990587_22990738 | 0.27 |
RP4-753D10.5 |
|
20760 |
0.15 |
| chr15_68376694_68376845 | 0.27 |
PIAS1 |
protein inhibitor of activated STAT, 1 |
29855 |
0.22 |
| chr19_18575570_18575881 | 0.26 |
ELL |
elongation factor RNA polymerase II |
11018 |
0.1 |
| chr19_39187756_39187907 | 0.26 |
CTB-186G2.4 |
|
3585 |
0.14 |
| chr1_31870789_31870940 | 0.26 |
SERINC2 |
serine incorporator 2 |
11548 |
0.15 |
| chr19_38423275_38423557 | 0.26 |
SIPA1L3 |
signal-induced proliferation-associated 1 like 3 |
25548 |
0.15 |
| chr3_58320540_58320691 | 0.26 |
PXK |
PX domain containing serine/threonine kinase |
1582 |
0.4 |
| chr1_33853388_33853681 | 0.26 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
12340 |
0.15 |
| chr6_35568601_35569436 | 0.26 |
ENSG00000212579 |
. |
50577 |
0.1 |
| chr10_3134024_3134295 | 0.26 |
PFKP |
phosphofructokinase, platelet |
12757 |
0.24 |
| chr16_17566923_17567074 | 0.26 |
XYLT1 |
xylosyltransferase I |
2260 |
0.48 |
| chr2_217051631_217052157 | 0.26 |
AC012513.6 |
|
72424 |
0.09 |
| chr12_6198255_6198797 | 0.26 |
ENSG00000240533 |
. |
6610 |
0.23 |
| chr12_113684408_113684559 | 0.26 |
TPCN1 |
two pore segment channel 1 |
2310 |
0.23 |
| chr13_80443621_80443990 | 0.26 |
NDFIP2 |
Nedd4 family interacting protein 2 |
388217 |
0.01 |
| chr5_75969089_75969240 | 0.26 |
F2R |
coagulation factor II (thrombin) receptor |
42704 |
0.11 |
| chr1_185703151_185703455 | 0.25 |
HMCN1 |
hemicentin 1 |
380 |
0.92 |
| chr1_234755566_234755985 | 0.25 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
10504 |
0.19 |
| chr15_96898066_96898266 | 0.25 |
AC087477.1 |
Uncharacterized protein |
6321 |
0.18 |
| chr12_124992848_124993269 | 0.25 |
NCOR2 |
nuclear receptor corepressor 2 |
9782 |
0.31 |
| chr5_171382567_171382718 | 0.25 |
FBXW11 |
F-box and WD repeat domain containing 11 |
22118 |
0.23 |
| chr1_59034555_59034706 | 0.25 |
ENSG00000264128 |
. |
3026 |
0.26 |
| chr6_49293968_49294177 | 0.25 |
ENSG00000252457 |
. |
18449 |
0.26 |
| chr16_4103086_4103844 | 0.25 |
RP11-462G12.4 |
|
21452 |
0.2 |
| chr12_78579294_78579445 | 0.25 |
NAV3 |
neuron navigator 3 |
13487 |
0.3 |
| chr1_201463290_201463773 | 0.25 |
CSRP1 |
cysteine and glycine-rich protein 1 |
2170 |
0.26 |
| chr19_18491035_18491248 | 0.25 |
GDF15 |
growth differentiation factor 15 |
1839 |
0.18 |
| chr5_124895396_124895838 | 0.25 |
ENSG00000222107 |
. |
208793 |
0.03 |
| chr16_83859962_83860113 | 0.25 |
HSBP1 |
heat shock factor binding protein 1 |
18443 |
0.15 |
| chr2_192530317_192530468 | 0.25 |
NABP1 |
nucleic acid binding protein 1 |
12470 |
0.29 |
| chr9_118403979_118404130 | 0.25 |
DEC1 |
deleted in esophageal cancer 1 |
499957 |
0.0 |
| chr2_159955686_159955837 | 0.25 |
ENSG00000202029 |
. |
72107 |
0.11 |
| chr9_108873656_108873902 | 0.25 |
TMEM38B |
transmembrane protein 38B |
389811 |
0.01 |
| chr5_123646330_123646481 | 0.25 |
RP11-436H11.2 |
|
418119 |
0.01 |
| chr12_89764903_89765288 | 0.25 |
DUSP6 |
dual specificity phosphatase 6 |
18047 |
0.24 |
| chr20_1796404_1796650 | 0.24 |
SIRPA |
signal-regulatory protein alpha |
78627 |
0.09 |
| chr11_10828028_10828469 | 0.24 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
761 |
0.52 |
| chr5_169577969_169578120 | 0.24 |
FOXI1 |
forkhead box I1 |
45127 |
0.16 |
| chr16_56404978_56405129 | 0.24 |
RP11-441F2.5 |
|
17617 |
0.13 |
| chr3_197043921_197044177 | 0.24 |
DLG1 |
discs, large homolog 1 (Drosophila) |
17878 |
0.15 |
| chr6_33930401_33930552 | 0.24 |
ENSG00000221697 |
. |
37352 |
0.18 |
| chr7_17246525_17246676 | 0.24 |
AC003075.4 |
|
74327 |
0.11 |
| chr6_56514356_56514539 | 0.24 |
DST |
dystonin |
6653 |
0.32 |
| chr18_43325991_43326142 | 0.24 |
RP11-116O18.3 |
|
1114 |
0.51 |
| chr4_74863945_74864096 | 0.24 |
CXCL5 |
chemokine (C-X-C motif) ligand 5 |
476 |
0.73 |
| chr1_65643796_65643947 | 0.24 |
AK4 |
adenylate kinase 4 |
29985 |
0.19 |
| chr3_46607636_46607787 | 0.24 |
LRRC2 |
leucine rich repeat containing 2 |
329 |
0.83 |
| chr4_157242892_157243172 | 0.24 |
ENSG00000221189 |
. |
25234 |
0.27 |
| chr7_41046808_41047238 | 0.24 |
AC005160.3 |
|
231866 |
0.02 |
| chr22_45106414_45106589 | 0.24 |
ARHGAP8 |
Rho GTPase activating protein 8 |
8119 |
0.19 |
| chr3_130693410_130693561 | 0.24 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
7253 |
0.18 |
| chr8_128919633_128920053 | 0.24 |
TMEM75 |
transmembrane protein 75 |
40748 |
0.15 |
| chr7_86783673_86784219 | 0.24 |
DMTF1 |
cyclin D binding myb-like transcription factor 1 |
1999 |
0.29 |
| chr2_216658765_216659109 | 0.24 |
ENSG00000212055 |
. |
84705 |
0.1 |
| chr9_107687183_107687334 | 0.24 |
RP11-217B7.2 |
|
2576 |
0.3 |
| chr3_87309504_87309716 | 0.24 |
POU1F1 |
POU class 1 homeobox 1 |
16002 |
0.21 |
| chr4_157860965_157861230 | 0.24 |
PDGFC |
platelet derived growth factor C |
30958 |
0.19 |
| chr1_77980530_77980774 | 0.24 |
AK5 |
adenylate kinase 5 |
17140 |
0.22 |
| chr22_30023302_30023509 | 0.24 |
NF2 |
neurofibromin 2 (merlin) |
23417 |
0.14 |
| chr17_45005909_45006060 | 0.24 |
GOSR2 |
golgi SNAP receptor complex member 2 |
5421 |
0.17 |
| chr17_60004372_60004770 | 0.24 |
INTS2 |
integrator complex subunit 2 |
216 |
0.94 |
| chr1_9498520_9498686 | 0.24 |
ENSG00000252956 |
. |
766 |
0.72 |
| chr12_104274321_104274475 | 0.24 |
RP11-650K20.3 |
Uncharacterized protein |
39169 |
0.1 |
| chr8_134353152_134353303 | 0.24 |
NDRG1 |
N-myc downstream regulated 1 |
38962 |
0.2 |
| chr2_45822706_45822857 | 0.23 |
SRBD1 |
S1 RNA binding domain 1 |
15639 |
0.21 |
| chr1_168051282_168051433 | 0.23 |
GPR161 |
G protein-coupled receptor 161 |
22810 |
0.21 |
| chr17_66449042_66449331 | 0.23 |
WIPI1 |
WD repeat domain, phosphoinositide interacting 1 |
4376 |
0.23 |
| chr10_60347392_60347543 | 0.23 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
74567 |
0.12 |
| chr7_17081811_17082046 | 0.23 |
AGR3 |
anterior gradient 3 |
160317 |
0.04 |
| chr6_170535437_170535588 | 0.23 |
RP5-1086L22.1 |
|
36145 |
0.15 |
| chr17_26673764_26673915 | 0.23 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
6407 |
0.07 |
| chrX_9755049_9755347 | 0.23 |
SHROOM2 |
shroom family member 2 |
702 |
0.51 |
| chr2_174055819_174055970 | 0.23 |
MLK7-AS1 |
MLK7 antisense RNA 1 |
16446 |
0.26 |
| chr2_216529775_216530114 | 0.23 |
ENSG00000212055 |
. |
213698 |
0.02 |
| chr3_100279906_100280202 | 0.23 |
GPR128 |
G protein-coupled receptor 128 |
48379 |
0.14 |
| chr8_129179760_129180073 | 0.23 |
ENSG00000221261 |
. |
17554 |
0.23 |
| chr7_79783795_79783946 | 0.23 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
18799 |
0.26 |
| chr2_46534465_46534616 | 0.23 |
EPAS1 |
endothelial PAS domain protein 1 |
9999 |
0.27 |
| chr5_95430378_95430529 | 0.23 |
ENSG00000207578 |
. |
15611 |
0.25 |
| chr5_10366297_10366564 | 0.23 |
MARCH6 |
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
12551 |
0.14 |
| chr6_71125955_71126106 | 0.23 |
RP11-462G2.2 |
|
2377 |
0.3 |
| chr6_4126077_4126228 | 0.23 |
RP3-400B16.4 |
|
9505 |
0.16 |
| chr12_8845562_8845807 | 0.23 |
RIMKLB |
ribosomal modification protein rimK-like family member B |
4089 |
0.19 |
| chr2_217342754_217342905 | 0.23 |
AC098820.4 |
|
9090 |
0.16 |
| chr21_27762100_27762398 | 0.23 |
AP001596.6 |
|
206 |
0.95 |
| chr1_85710395_85710781 | 0.23 |
C1orf52 |
chromosome 1 open reading frame 52 |
14728 |
0.13 |
| chr6_42192050_42192201 | 0.23 |
MRPS10 |
mitochondrial ribosomal protein S10 |
6522 |
0.19 |
| chr12_47353443_47353741 | 0.23 |
PCED1B |
PC-esterase domain containing 1B |
119794 |
0.06 |
| chr4_138965881_138966032 | 0.23 |
ENSG00000250033 |
. |
12662 |
0.32 |
| chr2_153361073_153361241 | 0.23 |
FMNL2 |
formin-like 2 |
114935 |
0.07 |
| chr1_28653212_28653398 | 0.23 |
MED18 |
mediator complex subunit 18 |
2244 |
0.26 |
| chr6_15391218_15391573 | 0.23 |
JARID2 |
jumonji, AT rich interactive domain 2 |
9694 |
0.25 |
| chr14_103838670_103838952 | 0.23 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
12918 |
0.15 |
| chr8_66629441_66629592 | 0.23 |
RP11-707M3.3 |
|
2503 |
0.31 |
| chr7_157145222_157145792 | 0.23 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
12992 |
0.24 |
| chr1_214596757_214596908 | 0.23 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
41314 |
0.18 |
| chr17_74525558_74526004 | 0.22 |
CYGB |
cytoglobin |
2366 |
0.14 |
| chr7_41548569_41548720 | 0.22 |
INHBA-AS1 |
INHBA antisense RNA 1 |
184870 |
0.03 |
| chr20_44443013_44443361 | 0.22 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
1542 |
0.2 |
| chr5_142096782_142096998 | 0.22 |
FGF1 |
fibroblast growth factor 1 (acidic) |
19279 |
0.22 |
| chr18_51550193_51550445 | 0.22 |
ENSG00000221058 |
. |
62707 |
0.15 |
| chr7_77495923_77496170 | 0.22 |
PHTF2 |
putative homeodomain transcription factor 2 |
26549 |
0.18 |
| chr7_134133604_134133901 | 0.22 |
AKR1B1 |
aldo-keto reductase family 1, member B1 (aldose reductase) |
10142 |
0.26 |
| chr5_149189745_149189896 | 0.22 |
PPARGC1B |
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
22758 |
0.18 |
| chr5_53075823_53075974 | 0.22 |
ENSG00000263710 |
. |
2630 |
0.4 |
| chr6_43065272_43065495 | 0.22 |
PTK7 |
protein tyrosine kinase 7 |
20831 |
0.09 |
| chr4_55147977_55148291 | 0.22 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
38392 |
0.21 |
| chr2_36603652_36603803 | 0.22 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
20113 |
0.27 |
| chr19_41768990_41769258 | 0.22 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
548 |
0.66 |
| chr2_48545707_48546468 | 0.22 |
FOXN2 |
forkhead box N2 |
4238 |
0.26 |
| chr14_65173299_65173599 | 0.22 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
2134 |
0.36 |
| chr1_200150874_200151025 | 0.22 |
ENSG00000221403 |
. |
36987 |
0.2 |
| chr15_86016825_86017700 | 0.22 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
70009 |
0.09 |
| chr12_109151621_109151772 | 0.22 |
ENSG00000200060 |
. |
19436 |
0.13 |
| chr2_68464884_68465350 | 0.22 |
ENSG00000216115 |
. |
8059 |
0.14 |
| chr12_63180443_63180611 | 0.22 |
ENSG00000200296 |
. |
64154 |
0.12 |
| chr3_182990531_182990717 | 0.22 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
7492 |
0.21 |
| chr1_210407365_210407516 | 0.22 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
48 |
0.97 |
| chr5_52023447_52023598 | 0.22 |
ITGA1 |
integrin, alpha 1 |
60208 |
0.12 |
| chr2_40261108_40261259 | 0.22 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
8307 |
0.32 |
| chr6_157719107_157719258 | 0.22 |
ENSG00000252609 |
. |
6751 |
0.25 |
| chr1_44030361_44030512 | 0.22 |
PTPRF |
protein tyrosine phosphatase, receptor type, F |
19689 |
0.18 |
| chr1_245249339_245249490 | 0.22 |
EFCAB2 |
EF-hand calcium binding domain 2 |
2656 |
0.26 |
| chr16_69051284_69051469 | 0.22 |
ENSG00000202497 |
. |
10318 |
0.2 |
| chr18_32230366_32230628 | 0.21 |
DTNA |
dystrobrevin, alpha |
29004 |
0.24 |
| chr3_119771367_119771672 | 0.21 |
GSK3B |
glycogen synthase kinase 3 beta |
40994 |
0.16 |
| chr9_80539292_80539443 | 0.21 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
101452 |
0.08 |
| chr17_28060383_28060541 | 0.21 |
RP11-82O19.1 |
|
27659 |
0.13 |
| chr14_78060038_78060870 | 0.21 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
3210 |
0.25 |
| chr12_120763372_120763602 | 0.21 |
PLA2G1B |
phospholipase A2, group IB (pancreas) |
409 |
0.75 |
| chr15_71118836_71119061 | 0.21 |
RP11-138H8.6 |
|
23161 |
0.14 |
| chr12_101382135_101382286 | 0.21 |
RP11-350G24.1 |
|
50662 |
0.14 |
| chr12_94171736_94171888 | 0.21 |
RP11-887P2.5 |
|
40213 |
0.14 |
| chr6_139697731_139697996 | 0.21 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
2106 |
0.42 |
| chr4_140719894_140720045 | 0.21 |
ENSG00000252233 |
. |
4382 |
0.3 |
| chr18_74197341_74197735 | 0.21 |
ZNF516 |
zinc finger protein 516 |
5197 |
0.19 |
| chr9_95177326_95177477 | 0.21 |
OMD |
osteomodulin |
9342 |
0.18 |
| chr3_54925868_54926132 | 0.21 |
CACNA2D3-AS1 |
CACNA2D3 antisense RNA 1 |
9282 |
0.27 |
| chr7_106064567_106064855 | 0.21 |
CTB-111H14.1 |
|
80574 |
0.1 |
| chr9_116357080_116357231 | 0.21 |
RGS3 |
regulator of G-protein signaling 3 |
1389 |
0.46 |
| chr8_68256392_68257141 | 0.21 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
854 |
0.75 |
| chr9_110812607_110812758 | 0.21 |
ENSG00000222459 |
. |
131423 |
0.05 |
| chr2_18806956_18807107 | 0.21 |
NT5C1B |
5'-nucleotidase, cytosolic IB |
36193 |
0.15 |
| chr8_24858429_24858620 | 0.21 |
CTD-2168K21.2 |
|
44389 |
0.16 |
| chr3_113513746_113513897 | 0.21 |
GRAMD1C |
GRAM domain containing 1C |
43444 |
0.13 |
| chr7_22214667_22214818 | 0.21 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
18785 |
0.27 |
| chr2_27942684_27942835 | 0.21 |
AC074091.13 |
Uncharacterized protein |
4160 |
0.18 |
| chr11_129509524_129509675 | 0.21 |
TMEM45B |
transmembrane protein 45B |
176115 |
0.03 |
| chr4_141019812_141020255 | 0.21 |
RP11-392B6.1 |
|
29136 |
0.21 |
| chr19_45901981_45902244 | 0.21 |
PPP1R13L |
protein phosphatase 1, regulatory subunit 13 like |
6184 |
0.12 |
| chr1_3112832_3112983 | 0.21 |
PRDM16 |
PR domain containing 16 |
47789 |
0.15 |
| chr20_45949788_45949944 | 0.21 |
AL031666.2 |
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
2620 |
0.19 |
| chr5_149422888_149423039 | 0.21 |
ENSG00000238369 |
. |
13948 |
0.14 |
| chr12_93178634_93178785 | 0.21 |
EEA1 |
early endosome antigen 1 |
36166 |
0.17 |
| chr20_43970628_43970800 | 0.21 |
SDC4 |
syndecan 4 |
6350 |
0.12 |
| chr15_67224295_67224446 | 0.21 |
SMAD3 |
SMAD family member 3 |
131731 |
0.05 |
| chr14_69548197_69548348 | 0.21 |
ENSG00000206768 |
. |
62398 |
0.11 |
| chr2_206635568_206635719 | 0.21 |
AC007362.3 |
|
6913 |
0.29 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.2 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.1 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 0.2 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.1 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.2 | GO:0045217 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0033133 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.0 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) |
| 0.0 | 0.1 | GO:1903224 | endodermal cell fate commitment(GO:0001711) endodermal cell fate specification(GO:0001714) endodermal cell differentiation(GO:0035987) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.0 | GO:0070977 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.0 | GO:0014848 | urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) sodium ion transmembrane transport(GO:0035725) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.2 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0031664 | regulation of lipopolysaccharide-mediated signaling pathway(GO:0031664) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.0 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0010714 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0003157 | endocardium development(GO:0003157) endocardium morphogenesis(GO:0003160) endocardium formation(GO:0060214) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0051900 | regulation of mitochondrial depolarization(GO:0051900) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.2 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.0 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.0 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.2 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.2 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |