Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
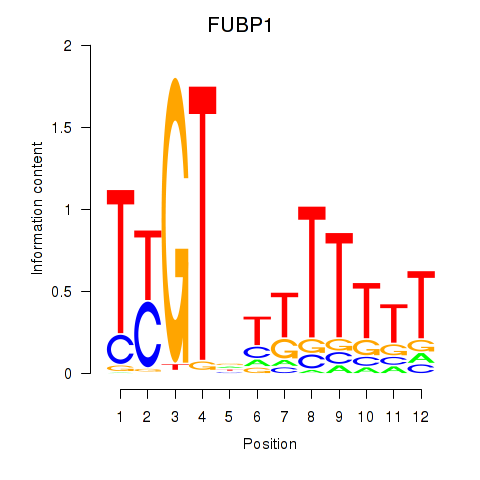
Results for FUBP1
Z-value: 1.30
Transcription factors associated with FUBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FUBP1
|
ENSG00000162613.12 | far upstream element binding protein 1 |
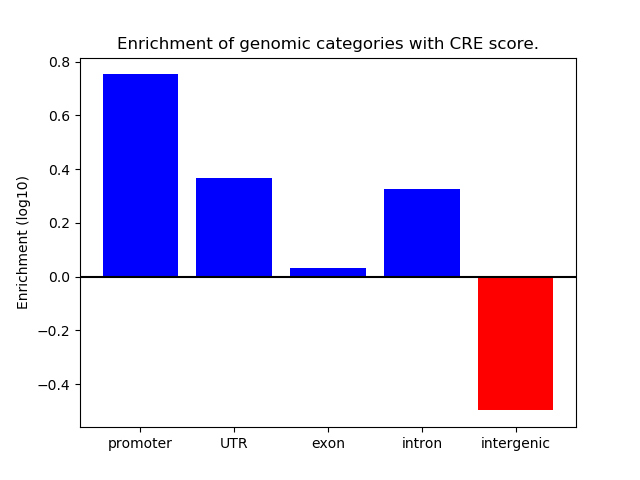
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_78416310_78416461 | FUBP1 | 28360 | 0.127943 | -0.73 | 2.7e-02 | Click! |
| chr1_78439235_78439436 | FUBP1 | 5410 | 0.170768 | -0.57 | 1.1e-01 | Click! |
| chr1_78440457_78440608 | FUBP1 | 4213 | 0.181598 | -0.48 | 1.9e-01 | Click! |
| chr1_78416706_78416961 | FUBP1 | 27912 | 0.128766 | -0.46 | 2.1e-01 | Click! |
| chr1_78436297_78436448 | FUBP1 | 8373 | 0.159883 | -0.43 | 2.5e-01 | Click! |
Activity of the FUBP1 motif across conditions
Conditions sorted by the z-value of the FUBP1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
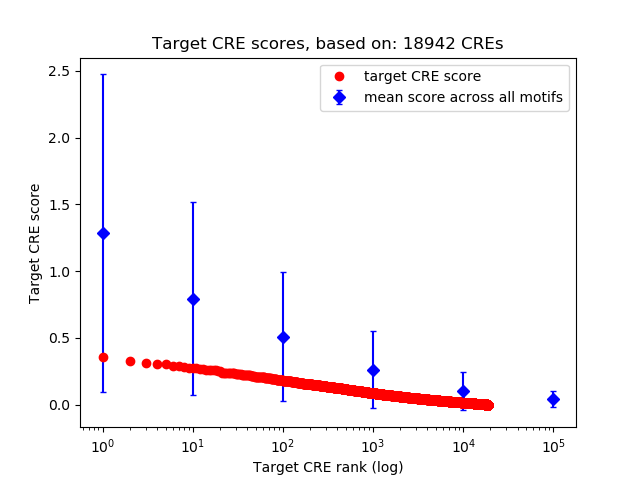
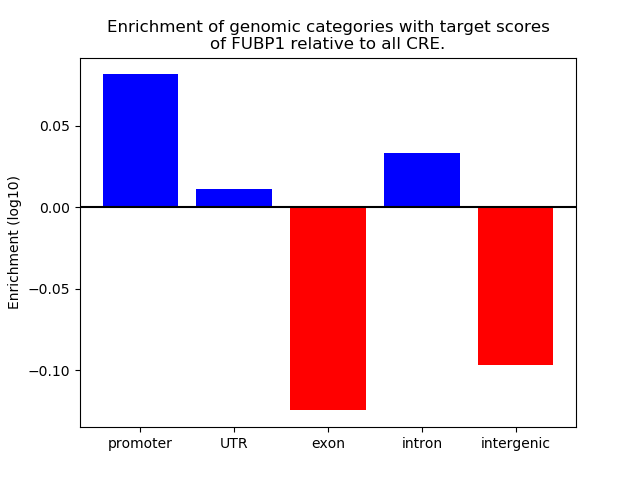
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_98717947_98718887 | 0.36 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
23225 |
0.2 |
| chr14_70180420_70180571 | 0.33 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
13122 |
0.24 |
| chr3_50410474_50410625 | 0.31 |
XXcos-LUCA11.4 |
|
5653 |
0.09 |
| chr16_30066961_30067112 | 0.31 |
ALDOA |
aldolase A, fructose-bisphosphate |
2581 |
0.11 |
| chr3_107763286_107763437 | 0.30 |
CD47 |
CD47 molecule |
13847 |
0.3 |
| chr2_191513228_191513379 | 0.29 |
NAB1 |
NGFI-A binding protein 1 (EGR1 binding protein 1) |
318 |
0.91 |
| chr21_46312526_46312824 | 0.29 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
17870 |
0.1 |
| chr14_98622440_98622591 | 0.28 |
ENSG00000222066 |
. |
175572 |
0.03 |
| chr12_111178547_111179341 | 0.28 |
PPP1CC |
protein phosphatase 1, catalytic subunit, gamma isozyme |
1700 |
0.39 |
| chr12_118768131_118768393 | 0.27 |
TAOK3 |
TAO kinase 3 |
28648 |
0.21 |
| chr6_103680064_103680234 | 0.27 |
ENSG00000202283 |
. |
350861 |
0.01 |
| chr8_142231323_142231474 | 0.27 |
SLC45A4 |
solute carrier family 45, member 4 |
7271 |
0.16 |
| chr19_10731127_10731278 | 0.27 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
4732 |
0.12 |
| chr6_119618545_119618696 | 0.26 |
ENSG00000200732 |
. |
29826 |
0.21 |
| chr1_40512120_40512273 | 0.26 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
5642 |
0.2 |
| chr21_16592051_16592291 | 0.26 |
NRIP1 |
nuclear receptor interacting protein 1 |
154850 |
0.04 |
| chr8_101227636_101227787 | 0.26 |
ENSG00000199667 |
. |
30345 |
0.14 |
| chrX_48859192_48859344 | 0.26 |
GRIPAP1 |
GRIP1 associated protein 1 |
593 |
0.56 |
| chr8_33412460_33412753 | 0.25 |
RNF122 |
ring finger protein 122 |
12037 |
0.13 |
| chr16_8818593_8818759 | 0.25 |
ABAT |
4-aminobutyrate aminotransferase |
4100 |
0.19 |
| chr5_133387222_133387373 | 0.24 |
VDAC1 |
voltage-dependent anion channel 1 |
46473 |
0.14 |
| chr15_91859349_91859500 | 0.24 |
SV2B |
synaptic vesicle glycoprotein 2B |
90324 |
0.09 |
| chr19_16695114_16695265 | 0.24 |
MED26 |
mediator complex subunit 26 |
3223 |
0.11 |
| chr10_33798752_33798923 | 0.24 |
NRP1 |
neuropilin 1 |
173647 |
0.03 |
| chr17_65420745_65420896 | 0.24 |
ENSG00000201547 |
. |
15823 |
0.13 |
| chrX_128756598_128756859 | 0.24 |
APLN |
apelin |
26014 |
0.21 |
| chr1_205599149_205600013 | 0.23 |
ELK4 |
ELK4, ETS-domain protein (SRF accessory protein 1) |
1509 |
0.35 |
| chrX_129254186_129254397 | 0.23 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
9600 |
0.2 |
| chr8_66588910_66589255 | 0.23 |
MTFR1 |
mitochondrial fission regulator 1 |
6866 |
0.22 |
| chr12_93984747_93985062 | 0.23 |
SOCS2 |
suppressor of cytokine signaling 2 |
16070 |
0.18 |
| chr6_14123211_14123362 | 0.23 |
CD83 |
CD83 molecule |
5414 |
0.28 |
| chr13_41929051_41929202 | 0.23 |
ENSG00000223280 |
. |
331 |
0.89 |
| chr19_16258305_16258456 | 0.23 |
HSH2D |
hematopoietic SH2 domain containing |
3834 |
0.15 |
| chr2_174818495_174818646 | 0.23 |
SP3 |
Sp3 transcription factor |
10377 |
0.31 |
| chr5_40685914_40686124 | 0.23 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
6419 |
0.22 |
| chrX_109275548_109275699 | 0.23 |
ENSG00000208013 |
. |
22934 |
0.19 |
| chr22_38795449_38796163 | 0.23 |
CSNK1E |
casein kinase 1, epsilon |
1279 |
0.34 |
| chr10_22540840_22540991 | 0.23 |
EBLN1 |
endogenous Bornavirus-like nucleoprotein 1 |
41965 |
0.14 |
| chr12_122898532_122898723 | 0.22 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
8489 |
0.22 |
| chr15_46162513_46162674 | 0.22 |
SQRDL |
sulfide quinone reductase-like (yeast) |
187855 |
0.03 |
| chr1_204553065_204553216 | 0.22 |
RP11-430C7.4 |
|
19023 |
0.14 |
| chr3_31975446_31975713 | 0.22 |
OSBPL10 |
oxysterol binding protein-like 10 |
47213 |
0.14 |
| chr1_83043773_83044235 | 0.22 |
LPHN2 |
latrophilin 2 |
598431 |
0.0 |
| chr20_58562413_58562564 | 0.22 |
CDH26 |
cadherin 26 |
1673 |
0.38 |
| chr12_54672984_54673135 | 0.22 |
CBX5 |
chromobox homolog 5 |
827 |
0.29 |
| chr3_182534933_182535084 | 0.21 |
ATP11B |
ATPase, class VI, type 11B |
19897 |
0.21 |
| chr16_78534160_78534433 | 0.21 |
RP11-264L1.4 |
|
6169 |
0.28 |
| chr12_24893513_24893767 | 0.21 |
ENSG00000240481 |
. |
33918 |
0.21 |
| chr19_47820020_47820171 | 0.21 |
C5AR1 |
complement component 5a receptor 1 |
6964 |
0.14 |
| chr1_100817300_100817579 | 0.21 |
CDC14A |
cell division cycle 14A |
149 |
0.96 |
| chr15_50346989_50347245 | 0.21 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
7533 |
0.26 |
| chr1_100819282_100819607 | 0.21 |
CDC14A |
cell division cycle 14A |
939 |
0.57 |
| chr17_79371033_79371247 | 0.21 |
RP11-1055B8.6 |
Uncharacterized protein |
1865 |
0.2 |
| chr5_148927186_148927463 | 0.21 |
CSNK1A1 |
casein kinase 1, alpha 1 |
2538 |
0.24 |
| chr2_201980395_201980546 | 0.21 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
357 |
0.82 |
| chr9_136933917_136934117 | 0.21 |
BRD3 |
bromodomain containing 3 |
360 |
0.86 |
| chr2_161289913_161290064 | 0.21 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
17299 |
0.2 |
| chr5_158267521_158267691 | 0.20 |
CTD-2363C16.1 |
|
142408 |
0.05 |
| chr2_234474872_234475097 | 0.20 |
USP40 |
ubiquitin specific peptidase 40 |
396 |
0.86 |
| chr21_36260768_36260938 | 0.20 |
RUNX1 |
runt-related transcription factor 1 |
134 |
0.98 |
| chr12_57077760_57078152 | 0.20 |
PTGES3 |
prostaglandin E synthase 3 (cytosolic) |
3990 |
0.14 |
| chr1_45668899_45669050 | 0.20 |
ZSWIM5 |
zinc finger, SWIM-type containing 5 |
3254 |
0.26 |
| chr4_86655280_86655719 | 0.20 |
ENSG00000266421 |
. |
11878 |
0.26 |
| chr21_47887630_47887781 | 0.20 |
DIP2A-IT1 |
DIP2A intronic transcript 1 (non-protein coding) |
5321 |
0.17 |
| chr5_37346370_37346521 | 0.20 |
ENSG00000251880 |
. |
19208 |
0.18 |
| chr20_62543850_62544001 | 0.20 |
ENSG00000199805 |
. |
3617 |
0.09 |
| chr9_123960312_123961002 | 0.20 |
RAB14 |
RAB14, member RAS oncogene family |
3490 |
0.19 |
| chr9_91047692_91047843 | 0.20 |
SPIN1 |
spindlin 1 |
44421 |
0.17 |
| chr9_100926805_100927131 | 0.20 |
CORO2A |
coronin, actin binding protein, 2A |
8198 |
0.21 |
| chr18_76738214_76738365 | 0.20 |
SALL3 |
spalt-like transcription factor 3 |
1986 |
0.46 |
| chr3_119806632_119806783 | 0.20 |
GSK3B |
glycogen synthase kinase 3 beta |
5806 |
0.22 |
| chr3_71751243_71751726 | 0.20 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
23042 |
0.2 |
| chr15_99398269_99398432 | 0.20 |
IGF1R |
insulin-like growth factor 1 receptor |
35220 |
0.16 |
| chr5_138638673_138638824 | 0.20 |
ENSG00000199545 |
. |
3207 |
0.14 |
| chr19_37339471_37339622 | 0.19 |
ZNF790 |
zinc finger protein 790 |
1669 |
0.24 |
| chr16_12910804_12910955 | 0.19 |
CPPED1 |
calcineurin-like phosphoesterase domain containing 1 |
13005 |
0.18 |
| chrX_15448302_15448453 | 0.19 |
BMX |
BMX non-receptor tyrosine kinase |
33992 |
0.15 |
| chr18_59856200_59856529 | 0.19 |
KIAA1468 |
KIAA1468 |
1840 |
0.31 |
| chr2_101946080_101946252 | 0.19 |
ENSG00000264857 |
. |
20254 |
0.16 |
| chr4_48374879_48375030 | 0.19 |
SLAIN2 |
SLAIN motif family, member 2 |
4964 |
0.28 |
| chr19_915577_915728 | 0.19 |
KISS1R |
KISS1 receptor |
1635 |
0.17 |
| chr15_69866363_69866514 | 0.19 |
ENSG00000207119 |
. |
115950 |
0.05 |
| chr12_120884573_120884899 | 0.19 |
GATC |
glutamyl-tRNA(Gln) amidotransferase, subunit C |
356 |
0.55 |
| chr16_87767946_87768097 | 0.19 |
RP11-278A23.2 |
|
13291 |
0.16 |
| chr13_28493501_28493652 | 0.19 |
PDX1 |
pancreatic and duodenal homeobox 1 |
581 |
0.66 |
| chr3_47170891_47171128 | 0.19 |
SETD2 |
SET domain containing 2 |
34057 |
0.13 |
| chr1_145661139_145661290 | 0.19 |
CD160 |
CD160 molecule |
45647 |
0.09 |
| chr19_19539763_19539914 | 0.19 |
GATAD2A |
GATA zinc finger domain containing 2A |
1502 |
0.3 |
| chr5_132288576_132288727 | 0.19 |
ENSG00000202417 |
. |
7617 |
0.15 |
| chr4_103741710_103742196 | 0.19 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
4746 |
0.17 |
| chr14_23386672_23386823 | 0.19 |
RBM23 |
RNA binding motif protein 23 |
18 |
0.93 |
| chrX_38661555_38661850 | 0.18 |
MID1IP1 |
MID1 interacting protein 1 |
995 |
0.5 |
| chr1_19794748_19794899 | 0.18 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
15756 |
0.16 |
| chr14_69401611_69401762 | 0.18 |
ACTN1 |
actinin, alpha 1 |
12579 |
0.23 |
| chr3_186674135_186674286 | 0.18 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
18535 |
0.2 |
| chr12_40627003_40627154 | 0.18 |
LRRK2 |
leucine-rich repeat kinase 2 |
8202 |
0.24 |
| chr19_1907912_1908063 | 0.18 |
ADAT3 |
adenosine deaminase, tRNA-specific 3 |
31 |
0.78 |
| chr13_33199531_33199682 | 0.18 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
39042 |
0.19 |
| chr10_115615104_115615263 | 0.18 |
NHLRC2 |
NHL repeat containing 2 |
763 |
0.48 |
| chr6_111790354_111790505 | 0.18 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
13985 |
0.19 |
| chr9_139158921_139159104 | 0.18 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
21325 |
0.15 |
| chr10_12632571_12632722 | 0.18 |
ENSG00000263584 |
. |
11894 |
0.27 |
| chrX_44884110_44884261 | 0.18 |
KDM6A |
lysine (K)-specific demethylase 6A |
34342 |
0.22 |
| chr16_84413717_84413987 | 0.18 |
ATP2C2 |
ATPase, Ca++ transporting, type 2C, member 2 |
11719 |
0.19 |
| chrX_119597210_119597803 | 0.18 |
LAMP2 |
lysosomal-associated membrane protein 2 |
5341 |
0.25 |
| chr19_50934673_50934858 | 0.18 |
MYBPC2 |
myosin binding protein C, fast type |
1395 |
0.23 |
| chr1_151196545_151196783 | 0.18 |
PIP5K1A |
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
10257 |
0.1 |
| chr2_134935532_134935768 | 0.18 |
ENSG00000263813 |
. |
50954 |
0.14 |
| chr13_48799322_48799473 | 0.18 |
ITM2B |
integral membrane protein 2B |
7897 |
0.27 |
| chr12_111807867_111808018 | 0.18 |
FAM109A |
family with sequence similarity 109, member A |
1017 |
0.54 |
| chr2_228362313_228362464 | 0.18 |
AGFG1 |
ArfGAP with FG repeats 1 |
10600 |
0.21 |
| chr1_27577181_27577392 | 0.18 |
ENSG00000252098 |
. |
4772 |
0.16 |
| chr3_188390892_188391164 | 0.18 |
ENSG00000207651 |
. |
15541 |
0.22 |
| chr11_1544049_1544200 | 0.18 |
MOB2 |
MOB kinase activator 2 |
36148 |
0.1 |
| chr1_171705490_171705830 | 0.18 |
VAMP4 |
vesicle-associated membrane protein 4 |
5542 |
0.2 |
| chr3_48477048_48477199 | 0.18 |
CCDC51 |
coiled-coil domain containing 51 |
2505 |
0.13 |
| chr7_101708245_101708396 | 0.18 |
CTA-357J21.1 |
|
32017 |
0.19 |
| chr5_43019074_43019523 | 0.18 |
CTD-2201E18.3 |
|
385 |
0.66 |
| chr13_30158034_30158185 | 0.18 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
2820 |
0.39 |
| chr2_182760548_182760802 | 0.18 |
SSFA2 |
sperm specific antigen 2 |
3752 |
0.25 |
| chr2_12959099_12960371 | 0.17 |
ENSG00000264370 |
. |
82242 |
0.11 |
| chr3_141045502_141045653 | 0.17 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
2522 |
0.33 |
| chr8_121738828_121738979 | 0.17 |
RP11-713M15.1 |
|
34590 |
0.2 |
| chr7_157679283_157679434 | 0.17 |
RP11-452C13.1 |
|
18128 |
0.22 |
| chr20_34327925_34328154 | 0.17 |
RBM39 |
RNA binding motif protein 39 |
720 |
0.55 |
| chr5_55964707_55964858 | 0.17 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
62723 |
0.12 |
| chr7_76851520_76851672 | 0.17 |
FGL2 |
fibrinogen-like 2 |
22453 |
0.19 |
| chr12_66698497_66698648 | 0.17 |
HELB |
helicase (DNA) B |
2247 |
0.27 |
| chr14_45627397_45627548 | 0.17 |
FANCM |
Fanconi anemia, complementation group M |
3510 |
0.21 |
| chr7_129500510_129500817 | 0.17 |
ENSG00000207691 |
. |
85809 |
0.06 |
| chr13_114119819_114119970 | 0.17 |
DCUN1D2-AS |
DCUN1D2 antisense RNA |
3322 |
0.17 |
| chr22_41683317_41683468 | 0.17 |
RANGAP1 |
Ran GTPase activating protein 1 |
1137 |
0.35 |
| chrX_44926778_44926929 | 0.17 |
KDM6A |
lysine (K)-specific demethylase 6A |
7551 |
0.3 |
| chr17_1082165_1082334 | 0.17 |
ABR |
active BCR-related |
829 |
0.6 |
| chr1_43411413_43411601 | 0.17 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
12993 |
0.18 |
| chr15_67356770_67357118 | 0.17 |
SMAD3 |
SMAD family member 3 |
843 |
0.73 |
| chr22_19073574_19073725 | 0.17 |
AC004471.9 |
|
35393 |
0.09 |
| chr12_10368361_10368512 | 0.17 |
RP11-656E20.5 |
|
1076 |
0.38 |
| chr3_16490080_16490231 | 0.17 |
RFTN1 |
raftlin, lipid raft linker 1 |
34217 |
0.19 |
| chr4_15681074_15681472 | 0.17 |
FBXL5 |
F-box and leucine-rich repeat protein 5 |
1904 |
0.29 |
| chr7_19155357_19155508 | 0.17 |
TWIST1 |
twist family bHLH transcription factor 1 |
1863 |
0.27 |
| chr4_143325972_143326254 | 0.17 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
26299 |
0.28 |
| chr19_46066687_46066838 | 0.17 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
21315 |
0.1 |
| chr11_75380064_75380270 | 0.17 |
MAP6 |
microtubule-associated protein 6 |
2 |
0.97 |
| chr2_216993992_216994143 | 0.17 |
XRCC5 |
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
19993 |
0.19 |
| chr15_63802358_63803207 | 0.17 |
USP3 |
ubiquitin specific peptidase 3 |
4863 |
0.26 |
| chr12_65091539_65091893 | 0.17 |
AC025262.1 |
Mesenchymal stem cell protein DSC96; Uncharacterized protein |
1387 |
0.36 |
| chr4_110512749_110512900 | 0.17 |
CCDC109B |
coiled-coil domain containing 109B |
30678 |
0.18 |
| chr22_40584317_40584468 | 0.17 |
TNRC6B |
trinucleotide repeat containing 6B |
10446 |
0.26 |
| chr12_69727071_69727222 | 0.17 |
LYZ |
lysozyme |
14975 |
0.17 |
| chr3_45053532_45053683 | 0.17 |
CLEC3B |
C-type lectin domain family 3, member B |
14068 |
0.15 |
| chrY_7035946_7036097 | 0.17 |
PRKY |
protein kinase, Y-linked, pseudogene |
106315 |
0.07 |
| chr6_119214221_119214767 | 0.17 |
ASF1A |
anti-silencing function 1A histone chaperone |
890 |
0.63 |
| chr17_65825111_65825409 | 0.16 |
BPTF |
bromodomain PHD finger transcription factor |
3002 |
0.23 |
| chr3_9797020_9797171 | 0.16 |
OGG1 |
8-oxoguanine DNA glycosylase |
3508 |
0.13 |
| chr17_54678795_54679086 | 0.16 |
NOG |
noggin |
7880 |
0.31 |
| chr8_67526501_67526877 | 0.16 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
1160 |
0.46 |
| chrX_131172208_131172403 | 0.16 |
MST4 |
Serine/threonine-protein kinase MST4 |
14669 |
0.21 |
| chr4_84035931_84036357 | 0.16 |
PLAC8 |
placenta-specific 8 |
189 |
0.96 |
| chr6_137579705_137579856 | 0.16 |
IFNGR1 |
interferon gamma receptor 1 |
39194 |
0.19 |
| chr6_156709882_156710033 | 0.16 |
ENSG00000212295 |
. |
10073 |
0.33 |
| chr6_149081739_149081986 | 0.16 |
UST |
uronyl-2-sulfotransferase |
13398 |
0.3 |
| chr15_86077549_86077700 | 0.16 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
9647 |
0.21 |
| chr15_77702308_77702459 | 0.16 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
10059 |
0.23 |
| chr20_48321244_48321782 | 0.16 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
8902 |
0.19 |
| chr22_42632196_42632347 | 0.16 |
TCF20 |
transcription factor 20 (AR1) |
20823 |
0.18 |
| chr8_42913895_42914749 | 0.16 |
FNTA |
farnesyltransferase, CAAX box, alpha |
227 |
0.91 |
| chr12_56609980_56610131 | 0.16 |
RNF41 |
ring finger protein 41 |
5430 |
0.07 |
| chr9_89562820_89562971 | 0.16 |
GAS1 |
growth arrest-specific 1 |
791 |
0.79 |
| chr9_20398844_20399100 | 0.16 |
ENSG00000263790 |
. |
12264 |
0.18 |
| chr11_73468535_73468693 | 0.16 |
RAB6A |
RAB6A, member RAS oncogene family |
2524 |
0.21 |
| chr6_12608393_12608818 | 0.16 |
PHACTR1 |
phosphatase and actin regulator 1 |
109288 |
0.07 |
| chr15_92414828_92415014 | 0.16 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
17570 |
0.25 |
| chr11_64529641_64529792 | 0.16 |
PYGM |
phosphorylase, glycogen, muscle |
1947 |
0.21 |
| chr2_101188537_101188688 | 0.16 |
PDCL3 |
phosducin-like 3 |
3195 |
0.21 |
| chr20_3996709_3996860 | 0.16 |
RNF24 |
ring finger protein 24 |
555 |
0.8 |
| chr2_62702609_62702952 | 0.16 |
ENSG00000241625 |
. |
15533 |
0.23 |
| chr9_135249138_135249289 | 0.16 |
SETX |
senataxin |
18841 |
0.17 |
| chr12_31470259_31470509 | 0.16 |
FAM60A |
family with sequence similarity 60, member A |
6712 |
0.16 |
| chr21_43149115_43149266 | 0.16 |
AP001615.9 |
|
10343 |
0.18 |
| chr2_68999011_68999272 | 0.16 |
ARHGAP25 |
Rho GTPase activating protein 25 |
2792 |
0.33 |
| chr17_49305755_49305970 | 0.16 |
MBTD1 |
mbt domain containing 1 |
3277 |
0.21 |
| chr4_169752762_169753076 | 0.16 |
PALLD |
palladin, cytoskeletal associated protein |
237 |
0.89 |
| chr16_14399831_14399982 | 0.16 |
ENSG00000207639 |
. |
2082 |
0.29 |
| chr7_27190679_27190886 | 0.16 |
HOXA-AS3 |
HOXA cluster antisense RNA 3 |
1111 |
0.21 |
| chrX_47929101_47929252 | 0.16 |
ZNF630 |
zinc finger protein 630 |
1307 |
0.34 |
| chr19_35758705_35758856 | 0.16 |
USF2 |
upstream transcription factor 2, c-fos interacting |
1101 |
0.29 |
| chr7_99739342_99739544 | 0.16 |
LAMTOR4 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
7087 |
0.06 |
| chr11_95781083_95781234 | 0.15 |
MTMR2 |
myotubularin related protein 2 |
123699 |
0.06 |
| chr5_72252681_72252978 | 0.15 |
FCHO2 |
FCH domain only 2 |
931 |
0.64 |
| chr10_131129546_131129832 | 0.15 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
135759 |
0.05 |
| chr15_93062736_93063031 | 0.15 |
C15orf32 |
chromosome 15 open reading frame 32 |
47732 |
0.15 |
| chr2_38595705_38595856 | 0.15 |
ATL2 |
atlastin GTPase 2 |
7586 |
0.3 |
| chr21_45079176_45079327 | 0.15 |
HSF2BP |
heat shock transcription factor 2 binding protein |
100 |
0.62 |
| chr17_38684308_38684459 | 0.15 |
TNS4 |
tensin 4 |
26534 |
0.13 |
| chr21_16582150_16582301 | 0.15 |
NRIP1 |
nuclear receptor interacting protein 1 |
144904 |
0.05 |
| chr12_77165937_77166088 | 0.15 |
ZDHHC17 |
zinc finger, DHHC-type containing 17 |
7659 |
0.3 |
| chr11_117199148_117199640 | 0.15 |
CEP164 |
centrosomal protein 164kDa |
575 |
0.46 |
| chr1_53739428_53739681 | 0.15 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
1781 |
0.2 |
| chr11_35061219_35061445 | 0.15 |
PDHX |
pyruvate dehydrogenase complex, component X |
62001 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0009188 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0046040 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.3 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.2 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.0 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.2 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.0 | GO:0051138 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.0 | GO:2000051 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.2 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.0 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.0 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.0 | GO:0060912 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.2 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) layer formation in cerebral cortex(GO:0021819) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0043558 | regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 | 0.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.1 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.0 | GO:0001991 | regulation of systemic arterial blood pressure by circulatory renin-angiotensin(GO:0001991) |
| 0.0 | 0.0 | GO:0050686 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.0 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.0 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0019348 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.0 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.0 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.0 | GO:0031943 | regulation of glucocorticoid metabolic process(GO:0031943) |
| 0.0 | 0.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0070193 | synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.3 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.1 | GO:0097526 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0044217 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0097525 | spliceosomal snRNP complex(GO:0097525) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.0 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:1903561 | extracellular organelle(GO:0043230) extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.0 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0004428 | obsolete inositol or phosphatidylinositol kinase activity(GO:0004428) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.0 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0015440 | oligopeptide-transporting ATPase activity(GO:0015421) peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.0 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.0 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.0 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |