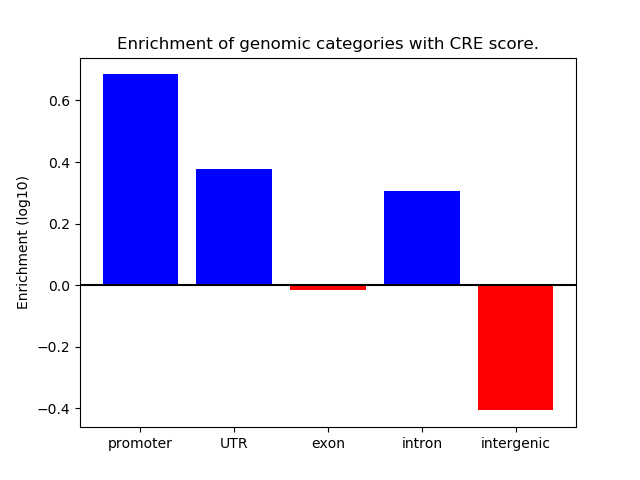
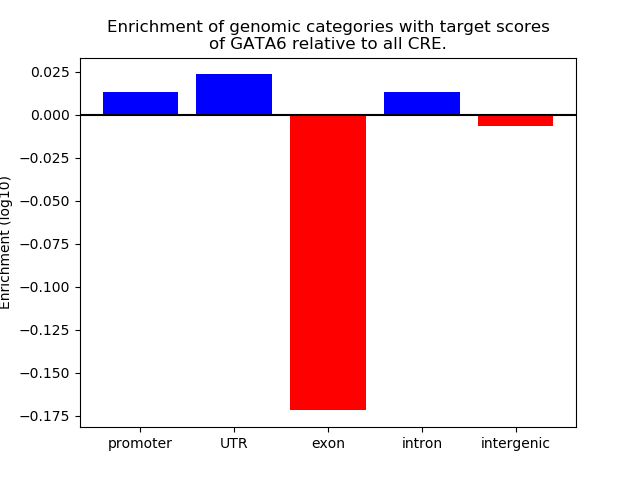
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
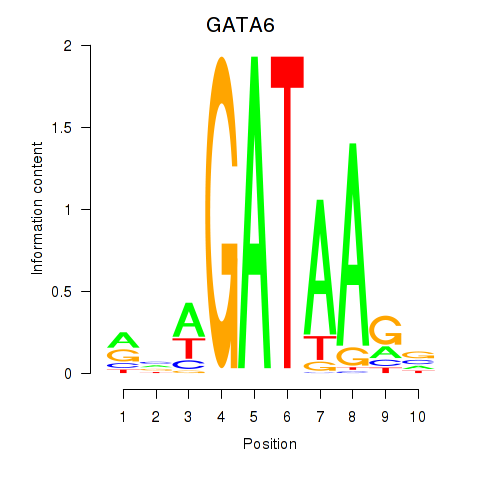
Results for GATA6
Z-value: 0.21
Transcription factors associated with GATA6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GATA6
|
ENSG00000141448.4 | GATA binding protein 6 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_19749352_19749503 | GATA6 | 23 | 0.968795 | 0.85 | 3.6e-03 | Click! |
| chr18_19750426_19750714 | GATA6 | 324 | 0.850451 | 0.76 | 1.7e-02 | Click! |
| chr18_19749074_19749225 | GATA6 | 255 | 0.892483 | 0.72 | 2.8e-02 | Click! |
| chr18_19745398_19745549 | GATA6 | 3931 | 0.172004 | 0.69 | 3.8e-02 | Click! |
| chr18_19749689_19749991 | GATA6 | 436 | 0.777502 | 0.67 | 4.9e-02 | Click! |
Activity of the GATA6 motif across conditions
Conditions sorted by the z-value of the GATA6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
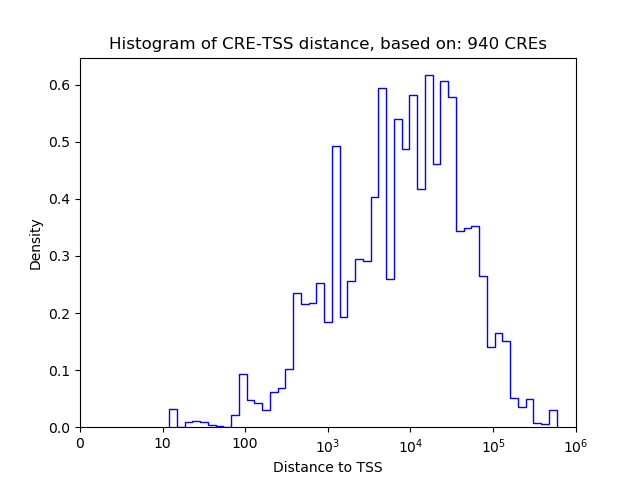
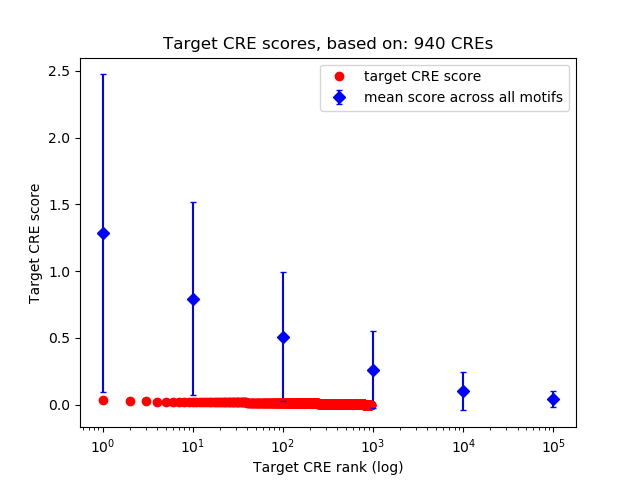
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_130628032_130628183 | 0.04 |
ENSG00000263979 |
. |
3667 |
0.1 |
| chr1_63260192_63260486 | 0.03 |
ATG4C |
autophagy related 4C, cysteine peptidase |
9435 |
0.28 |
| chr1_117528994_117529403 | 0.02 |
CD101 |
CD101 molecule |
15184 |
0.16 |
| chr9_100695698_100695849 | 0.02 |
C9orf156 |
chromosome 9 open reading frame 156 |
10921 |
0.14 |
| chr14_61870595_61870929 | 0.02 |
PRKCH |
protein kinase C, eta |
13350 |
0.24 |
| chr10_95171957_95172108 | 0.02 |
MYOF |
myoferlin |
69919 |
0.1 |
| chr7_50250289_50250673 | 0.02 |
AC020743.2 |
|
68062 |
0.11 |
| chr17_78676894_78677122 | 0.02 |
RP11-28G8.1 |
|
102424 |
0.06 |
| chr7_50353056_50353247 | 0.02 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
4833 |
0.32 |
| chr11_121460600_121460753 | 0.02 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
452 |
0.9 |
| chr1_230331179_230331330 | 0.02 |
RP5-956O18.2 |
|
72975 |
0.1 |
| chr6_159439729_159439953 | 0.02 |
RP1-111C20.4 |
|
7137 |
0.19 |
| chr1_239883809_239884265 | 0.02 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
1194 |
0.46 |
| chr15_52524118_52524315 | 0.02 |
GNB5 |
guanine nucleotide binding protein (G protein), beta 5 |
40650 |
0.12 |
| chr9_5842844_5842995 | 0.02 |
ERMP1 |
endoplasmic reticulum metallopeptidase 1 |
9802 |
0.2 |
| chr6_164386761_164386912 | 0.02 |
ENSG00000266128 |
. |
125243 |
0.06 |
| chr17_74812829_74813105 | 0.02 |
MFSD11 |
major facilitator superfamily domain containing 11 |
49444 |
0.08 |
| chr17_29638570_29638721 | 0.02 |
EVI2B |
ecotropic viral integration site 2B |
2457 |
0.2 |
| chr1_160537117_160537336 | 0.02 |
CD84 |
CD84 molecule |
12037 |
0.14 |
| chr12_70321695_70321901 | 0.02 |
MYRFL |
myelin regulatory factor-like |
1361 |
0.5 |
| chr8_97290954_97291105 | 0.02 |
PTDSS1 |
phosphatidylserine synthase 1 |
16391 |
0.15 |
| chr16_27244085_27244503 | 0.02 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
105 |
0.96 |
| chr19_16373633_16373875 | 0.02 |
CTD-2562J15.6 |
|
30632 |
0.12 |
| chr13_50552535_50552686 | 0.02 |
TRIM13 |
tripartite motif containing 13 |
17414 |
0.12 |
| chr1_26298239_26298465 | 0.02 |
ENSG00000207237 |
. |
7664 |
0.11 |
| chr20_57844647_57845067 | 0.02 |
EDN3 |
endothelin 3 |
30625 |
0.2 |
| chr8_97293950_97294101 | 0.02 |
PTDSS1 |
phosphatidylserine synthase 1 |
13395 |
0.16 |
| chr11_63648819_63649100 | 0.02 |
ENSG00000202089 |
. |
1198 |
0.38 |
| chr1_160605831_160606299 | 0.02 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
10746 |
0.16 |
| chr21_38597297_38597914 | 0.02 |
ENSG00000263969 |
. |
9699 |
0.16 |
| chr11_67178315_67178466 | 0.02 |
CARNS1 |
carnosine synthase 1 |
4759 |
0.07 |
| chr2_158767858_158768009 | 0.02 |
ENSG00000251980 |
. |
1351 |
0.44 |
| chr19_54898963_54899137 | 0.02 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
16885 |
0.09 |
| chr6_135509935_135510087 | 0.02 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
2855 |
0.25 |
| chr12_9177186_9177502 | 0.02 |
RP11-259O18.4 |
|
28182 |
0.13 |
| chr3_178983634_178983916 | 0.02 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
984 |
0.55 |
| chrX_70325744_70326037 | 0.02 |
CXorf65 |
chromosome X open reading frame 65 |
565 |
0.6 |
| chr7_38231978_38232129 | 0.02 |
STARD3NL |
STARD3 N-terminal like |
14056 |
0.3 |
| chr9_42926874_42927025 | 0.02 |
AC129778.2 |
|
67864 |
0.12 |
| chr2_198144726_198144925 | 0.02 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
22418 |
0.16 |
| chr17_32688274_32688552 | 0.02 |
CCL1 |
chemokine (C-C motif) ligand 1 |
1837 |
0.3 |
| chr21_47972699_47973025 | 0.02 |
ENSG00000272283 |
. |
17153 |
0.17 |
| chr6_159529160_159529540 | 0.02 |
RP11-13P5.2 |
|
59035 |
0.1 |
| chr11_114086673_114087539 | 0.02 |
NNMT |
nicotinamide N-methyltransferase |
41447 |
0.17 |
| chr9_5843022_5843173 | 0.02 |
ERMP1 |
endoplasmic reticulum metallopeptidase 1 |
9980 |
0.2 |
| chr2_112391271_112391480 | 0.02 |
ENSG00000266063 |
. |
137336 |
0.05 |
| chr2_98525890_98526041 | 0.02 |
TMEM131 |
transmembrane protein 131 |
2256 |
0.33 |
| chr3_32179771_32179931 | 0.02 |
GPD1L |
glycerol-3-phosphate dehydrogenase 1-like |
31660 |
0.15 |
| chr1_226921058_226921209 | 0.02 |
ITPKB |
inositol-trisphosphate 3-kinase B |
4026 |
0.28 |
| chr2_217630360_217630511 | 0.02 |
ENSG00000222832 |
. |
27888 |
0.15 |
| chr5_176356741_176356900 | 0.02 |
HK3 |
hexokinase 3 (white cell) |
30487 |
0.16 |
| chr1_66773813_66773964 | 0.02 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
22523 |
0.27 |
| chr1_206260544_206260695 | 0.02 |
ENSG00000252692 |
. |
474 |
0.79 |
| chr18_5293322_5293593 | 0.02 |
ZBTB14 |
zinc finger and BTB domain containing 14 |
1366 |
0.56 |
| chr9_82482306_82482544 | 0.02 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
160744 |
0.04 |
| chr22_40513481_40513654 | 0.02 |
TNRC6B |
trinucleotide repeat containing 6B |
60362 |
0.12 |
| chr6_137546259_137546508 | 0.02 |
IFNGR1 |
interferon gamma receptor 1 |
5797 |
0.28 |
| chr13_91796537_91796800 | 0.02 |
ENSG00000215417 |
. |
206191 |
0.03 |
| chr20_62739069_62739264 | 0.02 |
NPBWR2 |
neuropeptides B/W receptor 2 |
642 |
0.57 |
| chr3_10188290_10188441 | 0.02 |
VHL |
von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase |
4893 |
0.12 |
| chr2_169457921_169458072 | 0.02 |
ENSG00000199348 |
. |
5010 |
0.25 |
| chr9_67099892_67100043 | 0.02 |
ENSG00000265285 |
. |
5034 |
0.33 |
| chr7_77383911_77384062 | 0.02 |
RSBN1L |
round spermatid basic protein 1-like |
4926 |
0.25 |
| chr14_38677217_38677368 | 0.02 |
SSTR1 |
somatostatin receptor 1 |
88 |
0.97 |
| chr2_113428621_113428827 | 0.02 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
23445 |
0.17 |
| chr16_25104462_25104914 | 0.01 |
LCMT1 |
leucine carboxyl methyltransferase 1 |
18362 |
0.2 |
| chr21_36315540_36315813 | 0.01 |
RUNX1 |
runt-related transcription factor 1 |
53589 |
0.17 |
| chr9_95728001_95728291 | 0.01 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
1903 |
0.38 |
| chr11_112070521_112070755 | 0.01 |
BCO2 |
beta-carotene oxygenase 2 |
6441 |
0.11 |
| chr1_208056573_208056724 | 0.01 |
CD34 |
CD34 molecule |
7189 |
0.29 |
| chr2_98290201_98290458 | 0.01 |
LINC01125 |
long intergenic non-protein coding RNA 1125 |
462 |
0.76 |
| chrX_77038763_77038914 | 0.01 |
ATRX |
alpha thalassemia/mental retardation syndrome X-linked |
2864 |
0.32 |
| chr5_61636869_61637070 | 0.01 |
KIF2A |
kinesin heavy chain member 2A |
20164 |
0.22 |
| chr9_101878868_101879163 | 0.01 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
11042 |
0.2 |
| chr2_227702506_227702657 | 0.01 |
RHBDD1 |
rhomboid domain containing 1 |
317 |
0.91 |
| chr20_17491486_17491637 | 0.01 |
BFSP1 |
beaded filament structural protein 1, filensin |
20453 |
0.21 |
| chr10_2951549_2951726 | 0.01 |
PFKP |
phosphofructokinase, platelet |
156888 |
0.04 |
| chr3_160087726_160088066 | 0.01 |
IFT80 |
intraflagellar transport 80 homolog (Chlamydomonas) |
13548 |
0.15 |
| chr7_101497725_101497887 | 0.01 |
CTA-339C12.1 |
|
29797 |
0.18 |
| chr19_31841902_31842053 | 0.01 |
TSHZ3 |
teashirt zinc finger homeobox 3 |
1524 |
0.42 |
| chr2_231417375_231417526 | 0.01 |
AC010149.4 |
|
27198 |
0.15 |
| chr17_63169666_63169909 | 0.01 |
RGS9 |
regulator of G-protein signaling 9 |
36195 |
0.19 |
| chr9_93277885_93278036 | 0.01 |
DIRAS2 |
DIRAS family, GTP-binding RAS-like 2 |
127426 |
0.06 |
| chr8_128972530_128972700 | 0.01 |
ENSG00000221771 |
. |
264 |
0.92 |
| chr11_78165185_78165336 | 0.01 |
NARS2 |
asparaginyl-tRNA synthetase 2, mitochondrial (putative) |
10455 |
0.2 |
| chr14_70653199_70653350 | 0.01 |
RP11-486O13.2 |
|
566 |
0.77 |
| chr9_42574831_42574982 | 0.01 |
ENSG00000266353 |
. |
5067 |
0.31 |
| chr17_17048602_17048898 | 0.01 |
MPRIP |
myosin phosphatase Rho interacting protein |
8916 |
0.14 |
| chr11_114086418_114086569 | 0.01 |
NNMT |
nicotinamide N-methyltransferase |
42060 |
0.17 |
| chr2_217281812_217281963 | 0.01 |
SMARCAL1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
1219 |
0.41 |
| chr19_16474453_16474697 | 0.01 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
1811 |
0.29 |
| chr8_81961331_81961482 | 0.01 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
62897 |
0.14 |
| chr12_2901464_2901615 | 0.01 |
FKBP4 |
FK506 binding protein 4, 59kDa |
2580 |
0.18 |
| chr1_204329588_204329739 | 0.01 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
619 |
0.72 |
| chr7_32925349_32925561 | 0.01 |
KBTBD2 |
kelch repeat and BTB (POZ) domain containing 2 |
5310 |
0.24 |
| chr5_75810115_75810311 | 0.01 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
33021 |
0.2 |
| chr4_89526943_89527316 | 0.01 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
184 |
0.95 |
| chr6_26478781_26479098 | 0.01 |
BTN2A1 |
butyrophilin, subfamily 2, member A1 |
13484 |
0.12 |
| chr17_74117170_74117335 | 0.01 |
EXOC7 |
exocyst complex component 7 |
405 |
0.75 |
| chr12_125094240_125094476 | 0.01 |
NCOR2 |
nuclear receptor corepressor 2 |
42348 |
0.2 |
| chr1_40628068_40628453 | 0.01 |
RLF |
rearranged L-myc fusion |
1215 |
0.44 |
| chr7_50254432_50254583 | 0.01 |
AC020743.2 |
|
72088 |
0.11 |
| chr5_109192389_109192540 | 0.01 |
AC011366.3 |
Uncharacterized protein |
26419 |
0.26 |
| chr3_127202736_127202887 | 0.01 |
TPRA1 |
transmembrane protein, adipocyte asscociated 1 |
96469 |
0.07 |
| chr10_116365078_116365229 | 0.01 |
ENSG00000238577 |
. |
51116 |
0.15 |
| chr3_71754731_71754939 | 0.01 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
19691 |
0.21 |
| chr2_30482414_30482565 | 0.01 |
LBH |
limb bud and heart development |
27443 |
0.21 |
| chr8_126275118_126275439 | 0.01 |
ENSG00000242170 |
. |
7568 |
0.27 |
| chr11_6691639_6691994 | 0.01 |
MRPL17 |
mitochondrial ribosomal protein L17 |
12731 |
0.1 |
| chr12_58137526_58137677 | 0.01 |
TSPAN31 |
tetraspanin 31 |
1063 |
0.22 |
| chr1_113524173_113524422 | 0.01 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
24662 |
0.19 |
| chr10_4086226_4086442 | 0.01 |
KLF6 |
Kruppel-like factor 6 |
258861 |
0.02 |
| chr7_150179115_150179665 | 0.01 |
GIMAP8 |
GTPase, IMAP family member 8 |
31672 |
0.13 |
| chr10_31379930_31380081 | 0.01 |
ENSG00000263578 |
. |
25029 |
0.23 |
| chr13_100028628_100029061 | 0.01 |
ENSG00000207719 |
. |
20459 |
0.19 |
| chr15_66946404_66946592 | 0.01 |
SMAD6 |
SMAD family member 6 |
48068 |
0.12 |
| chr7_56019828_56020016 | 0.01 |
MRPS17 |
mitochondrial ribosomal protein S17 |
266 |
0.64 |
| chr13_28527067_28527218 | 0.01 |
ATP5EP2 |
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
7799 |
0.14 |
| chr7_36241182_36241445 | 0.01 |
AC007327.5 |
|
29403 |
0.18 |
| chr12_117488779_117488930 | 0.01 |
TESC |
tescalcin |
4218 |
0.3 |
| chr11_33925535_33925792 | 0.01 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
11827 |
0.21 |
| chr19_51629587_51629738 | 0.01 |
SIGLEC9 |
sialic acid binding Ig-like lectin 9 |
625 |
0.53 |
| chr16_75036571_75036722 | 0.01 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
451 |
0.84 |
| chr18_77190146_77190297 | 0.01 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
29829 |
0.2 |
| chr5_156297018_156297169 | 0.01 |
ENSG00000199468 |
. |
55952 |
0.13 |
| chr1_159055054_159055205 | 0.01 |
AIM2 |
absent in melanoma 2 |
8438 |
0.19 |
| chr14_50558921_50559072 | 0.01 |
C14orf183 |
chromosome 14 open reading frame 183 |
365 |
0.83 |
| chr9_21047360_21047561 | 0.01 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
15825 |
0.18 |
| chr9_126136303_126136518 | 0.01 |
CRB2 |
crumbs homolog 2 (Drosophila) |
5242 |
0.25 |
| chr8_99953508_99953739 | 0.01 |
STK3 |
serine/threonine kinase 3 |
1176 |
0.44 |
| chr6_135494903_135495054 | 0.01 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
7475 |
0.2 |
| chr22_38027238_38027450 | 0.01 |
SH3BP1 |
SH3-domain binding protein 1 |
8138 |
0.1 |
| chr1_114456887_114457038 | 0.01 |
AP4B1 |
adaptor-related protein complex 4, beta 1 subunit |
9139 |
0.12 |
| chr5_17218896_17219047 | 0.01 |
BASP1 |
brain abundant, membrane attached signal protein 1 |
1302 |
0.37 |
| chr2_75043564_75043871 | 0.01 |
HK2 |
hexokinase 2 |
17391 |
0.23 |
| chr16_76690596_76690747 | 0.01 |
RP11-58C22.1 |
Uncharacterized protein |
103357 |
0.08 |
| chr19_54858812_54858963 | 0.01 |
LILRA4 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4 |
8466 |
0.1 |
| chr10_97072027_97072178 | 0.01 |
PDLIM1 |
PDZ and LIM domain 1 |
21321 |
0.21 |
| chr6_35275141_35275407 | 0.01 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
2241 |
0.29 |
| chr12_104953946_104954167 | 0.01 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
28570 |
0.2 |
| chr19_48346448_48346599 | 0.01 |
TPRX2P |
tetra-peptide repeat homeobox 2 pseudogene |
8822 |
0.13 |
| chr2_99348262_99348529 | 0.01 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
806 |
0.7 |
| chr8_96224676_96225073 | 0.01 |
C8orf37 |
chromosome 8 open reading frame 37 |
56555 |
0.13 |
| chr3_16984516_16984667 | 0.01 |
ENSG00000264818 |
. |
9903 |
0.23 |
| chr17_58459442_58459593 | 0.01 |
USP32 |
ubiquitin specific peptidase 32 |
9819 |
0.21 |
| chr2_7061705_7061856 | 0.01 |
ENSG00000228203 |
. |
2940 |
0.24 |
| chr12_92798799_92799064 | 0.01 |
RP11-693J15.4 |
|
16376 |
0.18 |
| chr13_32562254_32562405 | 0.01 |
EEF1DP3 |
eukaryotic translation elongation factor 1 delta pseudogene 3 |
35612 |
0.17 |
| chr6_27599270_27599557 | 0.01 |
ENSG00000238648 |
. |
225 |
0.94 |
| chr8_8195680_8195853 | 0.01 |
SGK223 |
Tyrosine-protein kinase SgK223 |
43491 |
0.16 |
| chr20_42847628_42847998 | 0.01 |
OSER1 |
oxidative stress responsive serine-rich 1 |
8382 |
0.17 |
| chr20_37462715_37462866 | 0.01 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
28423 |
0.15 |
| chr6_137211346_137211580 | 0.01 |
SLC35D3 |
solute carrier family 35, member D3 |
31939 |
0.14 |
| chr10_95325510_95325661 | 0.01 |
FFAR4 |
free fatty acid receptor 4 |
837 |
0.58 |
| chr4_185285797_185285948 | 0.01 |
IRF2 |
interferon regulatory factor 2 |
53872 |
0.12 |
| chr14_22320921_22321072 | 0.01 |
ENSG00000222776 |
. |
72211 |
0.11 |
| chr1_42610621_42610772 | 0.01 |
GUCA2B |
guanylate cyclase activator 2B (uroguanylin) |
8396 |
0.21 |
| chr9_96362591_96362784 | 0.01 |
PHF2 |
PHD finger protein 2 |
23778 |
0.18 |
| chr5_77790229_77790380 | 0.01 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
54670 |
0.16 |
| chr5_156543326_156543684 | 0.01 |
HAVCR2 |
hepatitis A virus cellular receptor 2 |
6780 |
0.14 |
| chr3_27399507_27399712 | 0.01 |
ENSG00000221573 |
. |
4540 |
0.22 |
| chr11_128550867_128551052 | 0.01 |
RP11-744N12.3 |
|
5364 |
0.2 |
| chr9_117692590_117692826 | 0.01 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
11 |
0.99 |
| chr4_99529596_99529747 | 0.01 |
TSPAN5 |
tetraspanin 5 |
49115 |
0.15 |
| chr10_70832788_70833015 | 0.01 |
SRGN |
serglycin |
14973 |
0.18 |
| chr18_56390436_56390587 | 0.01 |
RP11-126O1.4 |
|
29451 |
0.14 |
| chr11_121325824_121326252 | 0.01 |
RP11-730K11.1 |
|
2316 |
0.33 |
| chr18_43808800_43809044 | 0.01 |
C18orf25 |
chromosome 18 open reading frame 25 |
54922 |
0.13 |
| chr4_76944504_76944655 | 0.01 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
71 |
0.95 |
| chr16_12661045_12661196 | 0.01 |
CTD-3037G24.4 |
|
9569 |
0.2 |
| chr9_124412947_124413098 | 0.01 |
DAB2IP |
DAB2 interacting protein |
851 |
0.7 |
| chr15_75447692_75447843 | 0.01 |
ENSG00000252722 |
. |
33760 |
0.11 |
| chr2_196378029_196378180 | 0.01 |
ENSG00000202206 |
. |
656 |
0.81 |
| chr2_138679588_138679739 | 0.01 |
HNMT |
histamine N-methyltransferase |
41927 |
0.22 |
| chr9_37301207_37301358 | 0.01 |
ENSG00000216070 |
. |
5181 |
0.23 |
| chr1_206292279_206292571 | 0.01 |
C1orf186 |
chromosome 1 open reading frame 186 |
3778 |
0.21 |
| chr5_58879154_58879305 | 0.01 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
2990 |
0.41 |
| chr16_67196662_67196813 | 0.01 |
HSF4 |
heat shock transcription factor 4 |
551 |
0.45 |
| chr7_93575157_93575308 | 0.01 |
AC006378.2 |
|
23522 |
0.14 |
| chr7_129545877_129546028 | 0.01 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
45215 |
0.1 |
| chr13_28002327_28002478 | 0.01 |
GTF3A |
general transcription factor IIIA |
1693 |
0.33 |
| chr5_135469095_135469294 | 0.01 |
SMAD5 |
SMAD family member 5 |
152 |
0.94 |
| chr5_173392661_173392812 | 0.01 |
C5orf47 |
chromosome 5 open reading frame 47 |
23426 |
0.21 |
| chr20_50087701_50087852 | 0.01 |
ENSG00000266761 |
. |
18262 |
0.24 |
| chr1_10474301_10474452 | 0.01 |
PGD |
phosphogluconate dehydrogenase |
14687 |
0.1 |
| chr1_39718531_39718682 | 0.01 |
RP11-420K8.1 |
|
4715 |
0.21 |
| chr8_27276778_27276929 | 0.01 |
PTK2B |
protein tyrosine kinase 2 beta |
11551 |
0.22 |
| chr1_25916924_25917087 | 0.01 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
26954 |
0.16 |
| chr15_78496258_78496409 | 0.01 |
ACSBG1 |
acyl-CoA synthetase bubblegum family member 1 |
3794 |
0.18 |
| chr4_90820392_90820543 | 0.01 |
MMRN1 |
multimerin 1 |
2667 |
0.36 |
| chr9_135936902_135937053 | 0.01 |
CEL |
carboxyl ester lipase |
388 |
0.79 |
| chr6_134953808_134954009 | 0.01 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
296391 |
0.01 |
| chr5_154191716_154191876 | 0.01 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
8015 |
0.17 |
| chr2_58957929_58958080 | 0.01 |
FANCL |
Fanconi anemia, complementation group L |
489497 |
0.01 |
| chr6_159480470_159480786 | 0.01 |
TAGAP |
T-cell activation RhoGTPase activating protein |
14444 |
0.19 |
| chr19_57171861_57172196 | 0.01 |
ZNF835 |
zinc finger protein 835 |
11100 |
0.14 |
| chr9_108462759_108462910 | 0.01 |
TMEM38B |
transmembrane protein 38B |
431 |
0.88 |
| chr6_136889409_136889560 | 0.01 |
MAP7 |
microtubule-associated protein 7 |
17527 |
0.15 |
| chr4_185744633_185744784 | 0.01 |
ACSL1 |
acyl-CoA synthetase long-chain family member 1 |
2480 |
0.27 |
| chr17_19141339_19141495 | 0.01 |
EPN2 |
epsin 2 |
685 |
0.56 |
Network of associatons between targets according to the STRING database.