Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
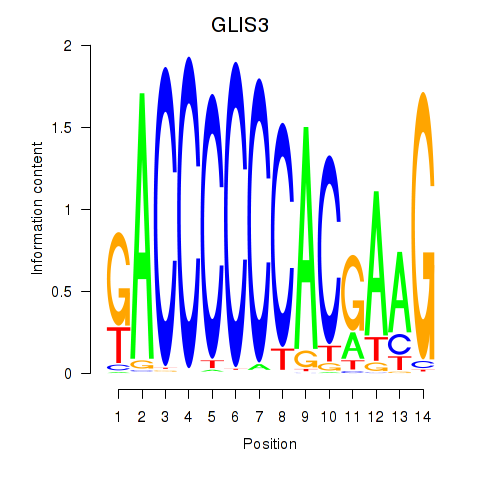
Results for GLIS3
Z-value: 1.31
Transcription factors associated with GLIS3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLIS3
|
ENSG00000107249.17 | GLIS family zinc finger 3 |
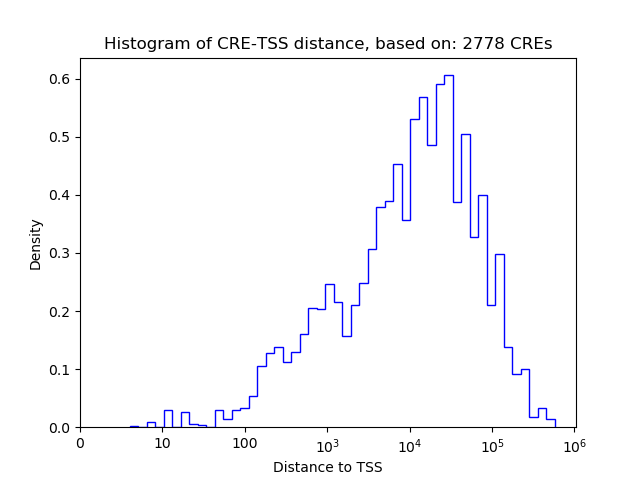
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_4292575_4292726 | GLIS3 | 5846 | 0.226245 | 0.94 | 2.1e-04 | Click! |
| chr9_4138215_4138366 | GLIS3 | 6903 | 0.284514 | 0.87 | 2.0e-03 | Click! |
| chr9_4225904_4226055 | GLIS3 | 72517 | 0.104295 | 0.85 | 3.4e-03 | Click! |
| chr9_4226493_4226644 | GLIS3 | 71928 | 0.105185 | 0.85 | 4.0e-03 | Click! |
| chr9_4263038_4263189 | GLIS3 | 35383 | 0.172344 | 0.85 | 4.0e-03 | Click! |
Activity of the GLIS3 motif across conditions
Conditions sorted by the z-value of the GLIS3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
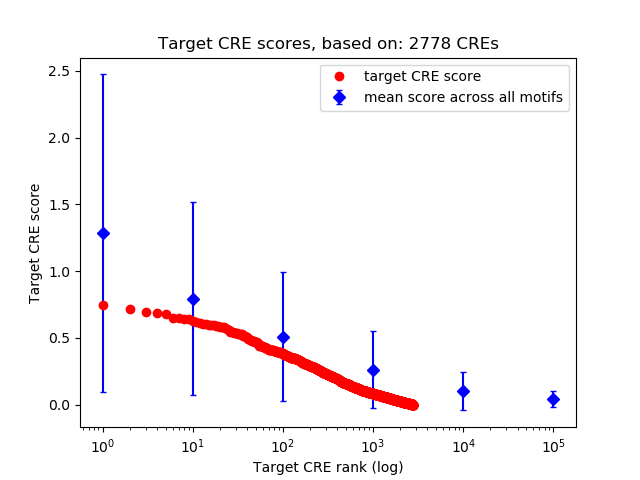
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_5504056_5504535 | 0.74 |
NET1 |
neuroepithelial cell transforming 1 |
15721 |
0.15 |
| chr18_57386294_57386563 | 0.71 |
CCBE1 |
collagen and calcium binding EGF domains 1 |
21816 |
0.15 |
| chr12_46886544_46886695 | 0.69 |
SLC38A2 |
solute carrier family 38, member 2 |
119969 |
0.06 |
| chr15_68469934_68470085 | 0.68 |
CALML4 |
calmodulin-like 4 |
22291 |
0.18 |
| chr8_49816316_49816467 | 0.68 |
SNAI2 |
snail family zinc finger 2 |
17597 |
0.29 |
| chr2_30888589_30888740 | 0.65 |
CAPN13 |
calpain 13 |
141647 |
0.05 |
| chr4_79220381_79220532 | 0.65 |
FRAS1 |
Fraser syndrome 1 |
44056 |
0.19 |
| chr14_69015594_69016105 | 0.65 |
CTD-2325P2.4 |
|
79313 |
0.1 |
| chr6_33641254_33641439 | 0.64 |
SBP1 |
SBP1; Uncharacterized protein |
22128 |
0.12 |
| chr7_71098288_71098439 | 0.63 |
ENSG00000199940 |
. |
52369 |
0.18 |
| chr14_105188331_105188482 | 0.62 |
ADSSL1 |
adenylosuccinate synthase like 1 |
2117 |
0.22 |
| chr9_38038631_38039019 | 0.61 |
SHB |
Src homology 2 domain containing adaptor protein B |
30383 |
0.2 |
| chr12_66393056_66393407 | 0.60 |
RP11-366L20.3 |
|
50972 |
0.12 |
| chr2_217163841_217163992 | 0.60 |
AC069155.1 |
|
4132 |
0.23 |
| chr7_116422296_116422447 | 0.60 |
MET |
met proto-oncogene |
5142 |
0.24 |
| chr12_66050364_66050557 | 0.60 |
HMGA2 |
high mobility group AT-hook 2 |
167451 |
0.03 |
| chr1_54801806_54802163 | 0.60 |
RP5-997D24.3 |
|
50906 |
0.12 |
| chr15_74344266_74344417 | 0.59 |
ENSG00000244612 |
. |
20698 |
0.13 |
| chr8_99014296_99014447 | 0.59 |
MATN2 |
matrilin 2 |
14401 |
0.15 |
| chr5_5019028_5019179 | 0.59 |
ENSG00000223007 |
. |
107056 |
0.07 |
| chr2_43202367_43202849 | 0.58 |
ENSG00000207087 |
. |
116024 |
0.06 |
| chr22_44845799_44845990 | 0.58 |
LDOC1L |
leucine zipper, down-regulated in cancer 1-like |
48284 |
0.16 |
| chr19_1417496_1417647 | 0.58 |
CTB-25B13.13 |
|
3671 |
0.09 |
| chr2_43201843_43202268 | 0.57 |
ENSG00000207087 |
. |
116577 |
0.06 |
| chr19_36009074_36009225 | 0.56 |
DMKN |
dermokine |
4589 |
0.11 |
| chr16_70785355_70785594 | 0.55 |
VAC14-AS1 |
VAC14 antisense RNA 1 |
3527 |
0.18 |
| chr13_110979517_110979668 | 0.54 |
COL4A2 |
collagen, type IV, alpha 2 |
19978 |
0.2 |
| chr1_42643557_42643708 | 0.54 |
FOXJ3 |
forkhead box J3 |
11848 |
0.19 |
| chr20_48643969_48644120 | 0.54 |
SNAI1 |
snail family zinc finger 1 |
44508 |
0.11 |
| chr16_67215208_67215359 | 0.54 |
KIAA0895L |
KIAA0895-like |
1318 |
0.19 |
| chr9_136580281_136580432 | 0.54 |
SARDH |
sarcosine dehydrogenase |
11533 |
0.2 |
| chr12_28543571_28543722 | 0.53 |
CCDC91 |
coiled-coil domain containing 91 |
59685 |
0.13 |
| chr17_77925591_77925742 | 0.53 |
TBC1D16 |
TBC1 domain family, member 16 |
146 |
0.96 |
| chr1_33723200_33724119 | 0.53 |
ZNF362 |
zinc finger protein 362 |
1485 |
0.38 |
| chr9_16031542_16031875 | 0.53 |
C9orf92 |
chromosome 9 open reading frame 92 |
184189 |
0.03 |
| chr20_48838016_48838310 | 0.52 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
30787 |
0.17 |
| chr6_109061444_109061595 | 0.52 |
FOXO3 |
forkhead box O3 |
83970 |
0.09 |
| chr17_70390304_70390455 | 0.52 |
ENSG00000200783 |
. |
269912 |
0.02 |
| chr5_67066501_67066705 | 0.51 |
ENSG00000223149 |
. |
196670 |
0.03 |
| chr13_30163335_30163486 | 0.51 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
2481 |
0.42 |
| chr3_99204006_99204160 | 0.49 |
ENSG00000266030 |
. |
118350 |
0.06 |
| chr1_203539117_203539268 | 0.49 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
56497 |
0.12 |
| chr6_52368912_52369217 | 0.49 |
TRAM2 |
translocation associated membrane protein 2 |
72649 |
0.1 |
| chr10_134224918_134226015 | 0.48 |
PWWP2B |
PWWP domain containing 2B |
14764 |
0.17 |
| chr15_39486358_39486509 | 0.48 |
C15orf54 |
chromosome 15 open reading frame 54 |
56452 |
0.15 |
| chr7_107635696_107635847 | 0.48 |
ENSG00000238297 |
. |
4656 |
0.19 |
| chr3_66474645_66474796 | 0.48 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
76636 |
0.11 |
| chr6_112366386_112366743 | 0.48 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
8711 |
0.21 |
| chr2_144713214_144713365 | 0.47 |
AC016910.1 |
|
18649 |
0.25 |
| chr3_120511291_120511442 | 0.47 |
GTF2E1 |
general transcription factor IIE, polypeptide 1, alpha 56kDa |
49775 |
0.14 |
| chr17_25571875_25572076 | 0.47 |
ENSG00000263583 |
. |
49047 |
0.15 |
| chr8_17519663_17519814 | 0.46 |
MTUS1 |
microtubule associated tumor suppressor 1 |
14100 |
0.17 |
| chr5_140887994_140888145 | 0.46 |
DIAPH1 |
diaphanous-related formin 1 |
17645 |
0.08 |
| chr17_71287002_71287642 | 0.45 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
20225 |
0.15 |
| chr1_225934809_225934960 | 0.44 |
ENSG00000223306 |
. |
5907 |
0.16 |
| chr11_119287635_119287786 | 0.44 |
RP11-334E6.12 |
|
951 |
0.43 |
| chr11_31991077_31991228 | 0.44 |
RP1-17K7.2 |
|
71642 |
0.09 |
| chr7_32626574_32626805 | 0.44 |
AVL9 |
AVL9 homolog (S. cerevisiase) |
43822 |
0.18 |
| chr1_27025127_27026141 | 0.44 |
ARID1A |
AT rich interactive domain 1A (SWI-like) |
2739 |
0.21 |
| chr12_53267484_53267947 | 0.44 |
KRT78 |
keratin 78 |
24839 |
0.11 |
| chr15_39006768_39007056 | 0.43 |
C15orf53 |
chromosome 15 open reading frame 53 |
18113 |
0.27 |
| chr10_52337444_52338204 | 0.43 |
ENSG00000251950 |
. |
7639 |
0.19 |
| chr6_121870706_121870857 | 0.43 |
ENSG00000201379 |
. |
7009 |
0.24 |
| chr13_110869599_110869750 | 0.43 |
COL4A2 |
collagen, type IV, alpha 2 |
88485 |
0.08 |
| chr3_151554670_151555319 | 0.42 |
AADAC |
arylacetamide deacetylase |
23133 |
0.18 |
| chr7_140042767_140042918 | 0.42 |
SLC37A3 |
solute carrier family 37, member 3 |
447 |
0.79 |
| chr5_33854965_33855116 | 0.42 |
ENSG00000201623 |
. |
33227 |
0.18 |
| chr14_96556907_96557058 | 0.42 |
C14orf132 |
chromosome 14 open reading frame 132 |
386 |
0.88 |
| chr5_145940287_145940438 | 0.42 |
CTB-99A3.1 |
|
2427 |
0.27 |
| chr16_4349012_4349227 | 0.41 |
GLIS2 |
GLIS family zinc finger 2 |
15643 |
0.13 |
| chr9_128640336_128640592 | 0.41 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
12914 |
0.3 |
| chr22_46933945_46934657 | 0.41 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
1234 |
0.46 |
| chr2_121673003_121673154 | 0.41 |
GLI2 |
GLI family zinc finger 2 |
118157 |
0.06 |
| chr3_52224733_52224962 | 0.41 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
7255 |
0.12 |
| chr14_55150516_55150667 | 0.41 |
SAMD4A |
sterile alpha motif domain containing 4A |
70960 |
0.12 |
| chr17_76953235_76953386 | 0.41 |
LGALS3BP |
lectin, galactoside-binding, soluble, 3 binding protein |
20095 |
0.13 |
| chr4_15481340_15481491 | 0.41 |
CC2D2A |
coiled-coil and C2 domain containing 2A |
568 |
0.78 |
| chr20_2748378_2748529 | 0.41 |
CPXM1 |
carboxypeptidase X (M14 family), member 1 |
32830 |
0.1 |
| chr10_29283078_29283229 | 0.40 |
ENSG00000199402 |
. |
119717 |
0.06 |
| chr14_55073033_55073184 | 0.40 |
SAMD4A |
sterile alpha motif domain containing 4A |
38471 |
0.18 |
| chr5_17235667_17236039 | 0.40 |
ENSG00000252908 |
. |
4867 |
0.21 |
| chr2_127852771_127853024 | 0.40 |
BIN1 |
bridging integrator 1 |
11680 |
0.25 |
| chr19_15321258_15321624 | 0.40 |
NOTCH3 |
notch 3 |
9649 |
0.16 |
| chrX_45570427_45570578 | 0.40 |
ENSG00000207870 |
. |
35192 |
0.19 |
| chr10_49798371_49798534 | 0.40 |
ARHGAP22 |
Rho GTPase activating protein 22 |
14545 |
0.21 |
| chr12_116820513_116820722 | 0.40 |
ENSG00000264037 |
. |
45506 |
0.17 |
| chr2_220898988_220899139 | 0.40 |
ENSG00000266518 |
. |
127777 |
0.05 |
| chr18_20945412_20945694 | 0.40 |
TMEM241 |
transmembrane protein 241 |
72264 |
0.09 |
| chr12_26266159_26266310 | 0.40 |
SSPN |
sarcospan |
8690 |
0.18 |
| chr21_39617669_39617820 | 0.40 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
10919 |
0.26 |
| chr6_7166165_7166316 | 0.39 |
RP11-405O10.2 |
|
17076 |
0.17 |
| chr9_90002642_90002793 | 0.39 |
DAPK1 |
death-associated protein kinase 1 |
109426 |
0.07 |
| chr3_52162372_52162604 | 0.39 |
POC1A |
POC1 centriolar protein A |
25932 |
0.12 |
| chr1_234858071_234858911 | 0.39 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
113220 |
0.06 |
| chr2_111223538_111223854 | 0.39 |
LIMS3 |
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein |
6697 |
0.18 |
| chr8_104145205_104145356 | 0.39 |
BAALC |
brain and acute leukemia, cytoplasmic |
7658 |
0.13 |
| chr17_2310691_2310842 | 0.39 |
MNT |
MAX network transcriptional repressor |
6354 |
0.12 |
| chr18_46449398_46450165 | 0.38 |
SMAD7 |
SMAD family member 7 |
25094 |
0.21 |
| chr8_95703738_95703889 | 0.38 |
ENSG00000199701 |
. |
22227 |
0.14 |
| chr10_121200841_121200992 | 0.38 |
GRK5 |
G protein-coupled receptor kinase 5 |
62967 |
0.1 |
| chr4_7888979_7889130 | 0.38 |
AFAP1 |
actin filament associated protein 1 |
15014 |
0.22 |
| chr22_20777294_20777549 | 0.38 |
ENSG00000207343 |
. |
6232 |
0.12 |
| chr17_45198047_45198198 | 0.38 |
ENSG00000221016 |
. |
2254 |
0.24 |
| chr6_43730965_43731135 | 0.38 |
VEGFA |
vascular endothelial growth factor A |
6900 |
0.18 |
| chr15_75639915_75640464 | 0.37 |
NEIL1 |
nei endonuclease VIII-like 1 (E. coli) |
12 |
0.96 |
| chr11_78798308_78798459 | 0.37 |
ENSG00000266550 |
. |
113529 |
0.07 |
| chr3_135615967_135616118 | 0.37 |
PPP2R3A |
protein phosphatase 2, regulatory subunit B'', alpha |
68473 |
0.13 |
| chr6_13395952_13396198 | 0.37 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
12294 |
0.21 |
| chr5_658132_658283 | 0.37 |
AC026740.1 |
Uncharacterized protein |
9552 |
0.15 |
| chr9_94137544_94137759 | 0.37 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
13456 |
0.27 |
| chr11_107566537_107566688 | 0.36 |
SLN |
sarcolipin |
16175 |
0.18 |
| chr16_2547240_2547391 | 0.36 |
TBC1D24 |
TBC1 domain family, member 24 |
1008 |
0.22 |
| chr22_28122999_28123150 | 0.36 |
RP11-375H17.1 |
|
10606 |
0.28 |
| chr21_28794694_28794845 | 0.36 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
455937 |
0.01 |
| chr20_57129971_57130122 | 0.36 |
APCDD1L |
adenomatosis polyposis coli down-regulated 1-like |
39859 |
0.14 |
| chr9_139844616_139845130 | 0.36 |
LCN12 |
lipocalin 12 |
1907 |
0.13 |
| chr17_7791584_7792259 | 0.36 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
188 |
0.87 |
| chr17_80194576_80195120 | 0.36 |
SLC16A3 |
solute carrier family 16 (monocarboxylate transporter), member 3 |
198 |
0.89 |
| chr1_202545648_202545889 | 0.36 |
RP11-569A11.1 |
|
27628 |
0.17 |
| chr12_12824967_12825118 | 0.36 |
GPR19 |
G protein-coupled receptor 19 |
12413 |
0.17 |
| chr2_120011300_120011451 | 0.35 |
STEAP3-AS1 |
STEAP3 antisense RNA 1 |
4728 |
0.22 |
| chr9_35526524_35526675 | 0.35 |
RUSC2 |
RUN and SH3 domain containing 2 |
12030 |
0.15 |
| chrX_69697338_69697582 | 0.35 |
DLG3-AS1 |
DLG3 antisense RNA 1 |
21616 |
0.14 |
| chr17_79486044_79486605 | 0.35 |
RP13-766D20.2 |
|
258 |
0.83 |
| chr7_137564480_137564636 | 0.35 |
DGKI |
diacylglycerol kinase, iota |
32720 |
0.18 |
| chr5_131604535_131604686 | 0.35 |
PDLIM4 |
PDZ and LIM domain 4 |
7468 |
0.16 |
| chr2_204206546_204206697 | 0.35 |
RP11-363J17.1 |
|
12672 |
0.2 |
| chr4_85888768_85889045 | 0.35 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
1362 |
0.57 |
| chr1_178027068_178027219 | 0.35 |
RASAL2 |
RAS protein activator like 2 |
35721 |
0.22 |
| chr21_44142964_44143115 | 0.35 |
AP001627.1 |
|
18829 |
0.2 |
| chr1_32123952_32124158 | 0.35 |
RP11-73M7.6 |
|
1196 |
0.35 |
| chr9_116928754_116928905 | 0.35 |
COL27A1 |
collagen, type XXVII, alpha 1 |
1166 |
0.54 |
| chr7_155137025_155137560 | 0.35 |
BLACE |
B-cell acute lymphoblastic leukemia expressed |
23337 |
0.16 |
| chr16_75057155_75057343 | 0.35 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
21054 |
0.18 |
| chr1_17914051_17914202 | 0.35 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
785 |
0.75 |
| chr15_67096793_67096944 | 0.35 |
SMAD6 |
SMAD family member 6 |
92833 |
0.08 |
| chr1_119822636_119822787 | 0.35 |
ENSG00000238679 |
. |
21047 |
0.19 |
| chr17_27459777_27460175 | 0.35 |
MYO18A |
myosin XVIIIA |
7460 |
0.15 |
| chr20_3657916_3658067 | 0.35 |
ADAM33 |
ADAM metallopeptidase domain 33 |
4759 |
0.17 |
| chr14_103984756_103984907 | 0.35 |
CKB |
creatine kinase, brain |
2607 |
0.14 |
| chr9_127027946_127028319 | 0.34 |
RP11-121A14.3 |
|
2977 |
0.22 |
| chr6_136151504_136151655 | 0.34 |
PDE7B |
phosphodiesterase 7B |
21255 |
0.25 |
| chr2_100171123_100171274 | 0.34 |
AFF3 |
AF4/FMR2 family, member 3 |
23634 |
0.19 |
| chr8_145017107_145017820 | 0.34 |
PLEC |
plectin |
663 |
0.52 |
| chr6_43243469_43244256 | 0.34 |
SLC22A7 |
solute carrier family 22 (organic anion transporter), member 7 |
21874 |
0.11 |
| chr17_35475957_35476108 | 0.34 |
ACACA |
acetyl-CoA carboxylase alpha |
30114 |
0.16 |
| chr2_20018811_20018962 | 0.33 |
TTC32 |
tetratricopeptide repeat domain 32 |
82516 |
0.1 |
| chr10_127916657_127916808 | 0.33 |
ENSG00000222740 |
. |
82581 |
0.1 |
| chr1_67142330_67142481 | 0.33 |
AL139147.1 |
Uncharacterized protein |
305 |
0.89 |
| chr9_140352686_140353315 | 0.33 |
NSMF |
NMDA receptor synaptonuclear signaling and neuronal migration factor |
754 |
0.52 |
| chr17_47928909_47929203 | 0.33 |
TAC4 |
tachykinin 4 (hemokinin) |
3677 |
0.2 |
| chr4_2061713_2062092 | 0.33 |
NAT8L |
N-acetyltransferase 8-like (GCN5-related, putative) |
363 |
0.84 |
| chr17_38469856_38470102 | 0.33 |
RARA |
retinoic acid receptor, alpha |
4533 |
0.14 |
| chr6_4733457_4733608 | 0.33 |
CDYL |
chromodomain protein, Y-like |
27139 |
0.2 |
| chr17_79438771_79438922 | 0.33 |
RP11-1055B8.8 |
|
9883 |
0.1 |
| chr2_113999292_113999443 | 0.33 |
PAX8 |
paired box 8 |
107 |
0.93 |
| chr2_218867800_218867951 | 0.33 |
TNS1 |
tensin 1 |
157 |
0.96 |
| chr20_43729390_43729742 | 0.32 |
KCNS1 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
184 |
0.93 |
| chr2_43504294_43504761 | 0.32 |
AC010883.5 |
|
47815 |
0.15 |
| chr19_1745168_1745319 | 0.32 |
ONECUT3 |
one cut homeobox 3 |
7129 |
0.14 |
| chr12_122467797_122467948 | 0.32 |
BCL7A |
B-cell CLL/lymphoma 7A |
8080 |
0.22 |
| chr9_35604415_35605122 | 0.32 |
TESK1 |
testis-specific kinase 1 |
599 |
0.54 |
| chr9_115608892_115609043 | 0.32 |
SNX30 |
sorting nexin family member 30 |
8102 |
0.21 |
| chr1_201420230_201420616 | 0.31 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
17889 |
0.14 |
| chr16_54169535_54169686 | 0.31 |
RP11-357N13.1 |
|
81115 |
0.09 |
| chr12_105762537_105762688 | 0.31 |
C12orf75 |
chromosome 12 open reading frame 75 |
37966 |
0.15 |
| chr17_34053980_34054131 | 0.31 |
RASL10B |
RAS-like, family 10, member B |
4613 |
0.13 |
| chr1_204857824_204857975 | 0.31 |
ENSG00000252946 |
. |
16047 |
0.2 |
| chr1_156634093_156634737 | 0.31 |
RP11-284F21.7 |
|
3199 |
0.12 |
| chr14_55120909_55121243 | 0.31 |
SAMD4A |
sterile alpha motif domain containing 4A |
86439 |
0.09 |
| chr9_137221477_137221979 | 0.31 |
RXRA |
retinoid X receptor, alpha |
3302 |
0.32 |
| chr11_94524719_94524870 | 0.31 |
AMOTL1 |
angiomotin like 1 |
23257 |
0.21 |
| chr2_16832198_16832526 | 0.31 |
FAM49A |
family with sequence similarity 49, member A |
14734 |
0.3 |
| chr16_58059108_58059507 | 0.31 |
MMP15 |
matrix metallopeptidase 15 (membrane-inserted) |
163 |
0.93 |
| chr2_109955799_109955950 | 0.31 |
ENSG00000265965 |
. |
25793 |
0.26 |
| chr9_16922088_16922239 | 0.31 |
BNC2 |
basonuclin 2 |
51322 |
0.18 |
| chr5_149939244_149939395 | 0.31 |
ENSG00000239191 |
. |
16068 |
0.16 |
| chr1_95189238_95189522 | 0.31 |
ENSG00000263526 |
. |
22076 |
0.24 |
| chr10_114794069_114794293 | 0.31 |
RP11-57H14.2 |
|
82547 |
0.09 |
| chr2_88202897_88203048 | 0.31 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
77501 |
0.09 |
| chr1_15059960_15060165 | 0.31 |
KAZN |
kazrin, periplakin interacting protein |
134849 |
0.05 |
| chr15_77775886_77776037 | 0.30 |
HMG20A |
high mobility group 20A |
5326 |
0.28 |
| chr9_130301500_130301651 | 0.30 |
FAM129B |
family with sequence similarity 129, member B |
29792 |
0.13 |
| chr9_94476107_94476273 | 0.30 |
ENSG00000266855 |
. |
77657 |
0.12 |
| chr16_75272514_75272665 | 0.30 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
4011 |
0.16 |
| chr3_11237382_11237744 | 0.30 |
HRH1 |
histamine receptor H1 |
30154 |
0.22 |
| chr16_72972999_72973226 | 0.30 |
ENSG00000221799 |
. |
45644 |
0.15 |
| chr17_14693509_14693957 | 0.30 |
ENSG00000238806 |
. |
447467 |
0.01 |
| chr15_69446958_69447109 | 0.30 |
GLCE |
glucuronic acid epimerase |
5940 |
0.2 |
| chr1_229118887_229119229 | 0.30 |
RP5-1061H20.5 |
|
244251 |
0.01 |
| chr4_189329009_189329160 | 0.30 |
RP11-366H4.3 |
|
264551 |
0.02 |
| chr1_47109437_47109588 | 0.30 |
ATPAF1 |
ATP synthase mitochondrial F1 complex assembly factor 1 |
1416 |
0.33 |
| chr6_85514798_85514949 | 0.30 |
TBX18 |
T-box 18 |
40636 |
0.21 |
| chr5_95594212_95594363 | 0.30 |
ENSG00000206997 |
. |
48356 |
0.17 |
| chr11_126417501_126417652 | 0.30 |
KIRREL3-AS1 |
KIRREL3 antisense RNA 1 |
3734 |
0.31 |
| chr5_141700206_141700587 | 0.29 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
3351 |
0.28 |
| chr16_85244563_85244846 | 0.29 |
CTC-786C10.1 |
|
39822 |
0.17 |
| chr15_80781999_80782170 | 0.29 |
RP11-210M15.1 |
|
44591 |
0.13 |
| chr11_89597443_89597867 | 0.29 |
TRIM64B |
tripartite motif containing 64B |
11530 |
0.22 |
| chrX_149428232_149428383 | 0.29 |
ENSG00000252454 |
. |
32068 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 0.3 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.2 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.2 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.4 | GO:0032069 | regulation of nuclease activity(GO:0032069) |
| 0.0 | 0.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.2 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.2 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.0 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.3 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.1 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.0 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0060768 | regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.1 | GO:0045628 | regulation of T-helper 2 cell differentiation(GO:0045628) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0031272 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0060179 | male courtship behavior(GO:0008049) male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.0 | GO:0034123 | positive regulation of toll-like receptor signaling pathway(GO:0034123) |
| 0.0 | 0.0 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.0 | GO:0035751 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.0 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0032432 | actin filament bundle(GO:0032432) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.2 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.3 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.0 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.0 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |