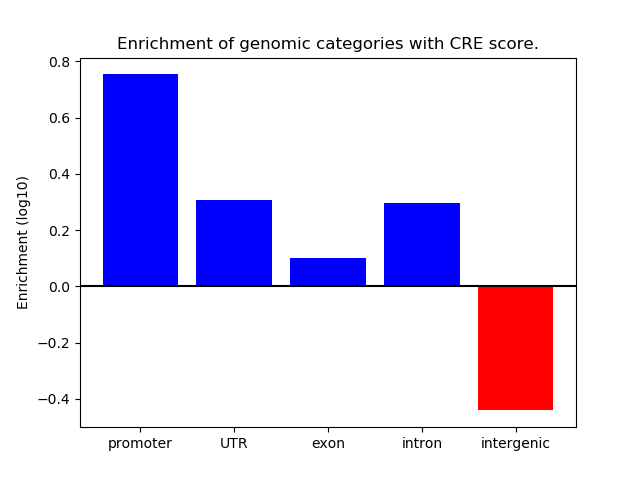
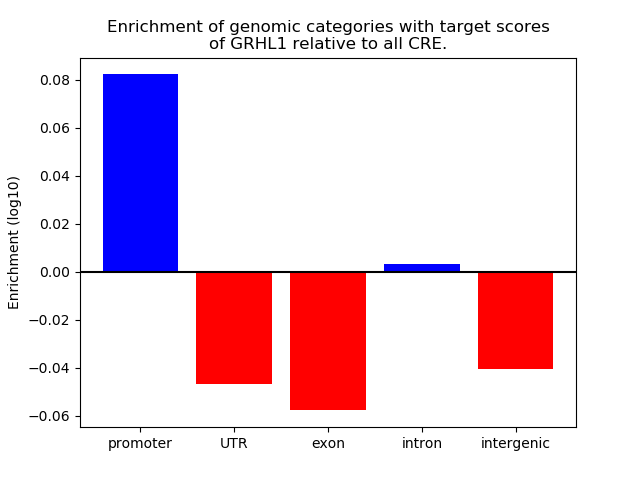
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
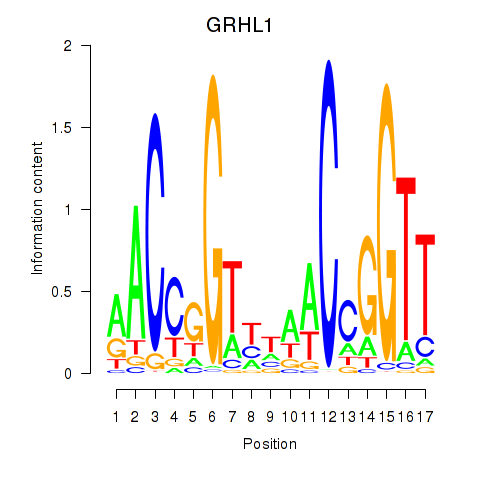
Results for GRHL1
Z-value: 2.41
Transcription factors associated with GRHL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GRHL1
|
ENSG00000134317.13 | grainyhead like transcription factor 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_10120083_10120407 | GRHL1 | 15189 | 0.161474 | -0.72 | 2.8e-02 | Click! |
| chr2_10106881_10107032 | GRHL1 | 15129 | 0.174178 | -0.68 | 4.5e-02 | Click! |
| chr2_10122799_10122950 | GRHL1 | 12560 | 0.163663 | -0.63 | 7.0e-02 | Click! |
| chr2_10109337_10109488 | GRHL1 | 17585 | 0.167183 | -0.56 | 1.2e-01 | Click! |
| chr2_10081034_10081185 | GRHL1 | 4232 | 0.239220 | 0.55 | 1.2e-01 | Click! |
Activity of the GRHL1 motif across conditions
Conditions sorted by the z-value of the GRHL1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
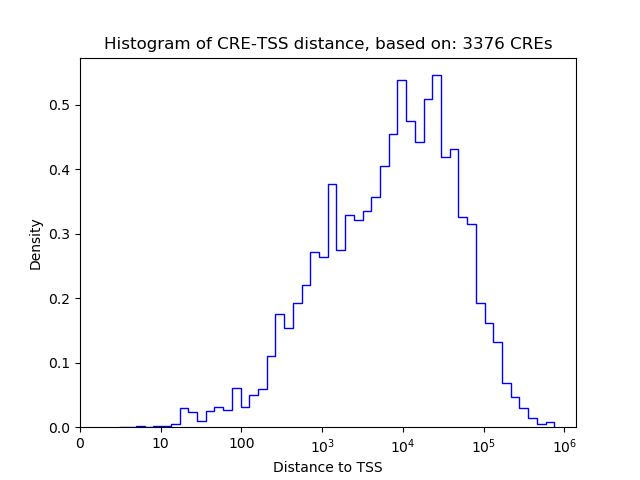
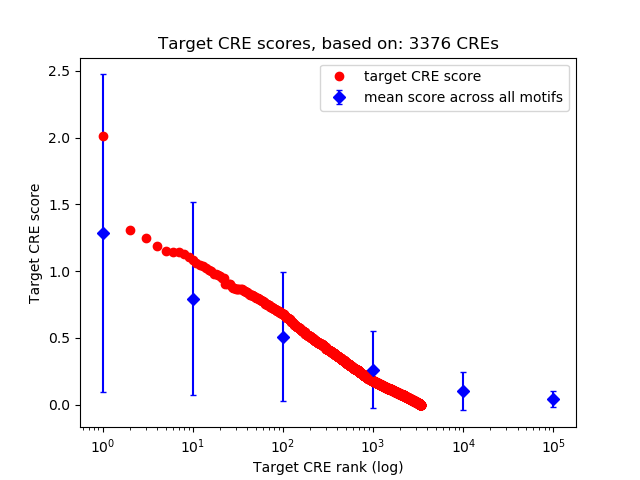
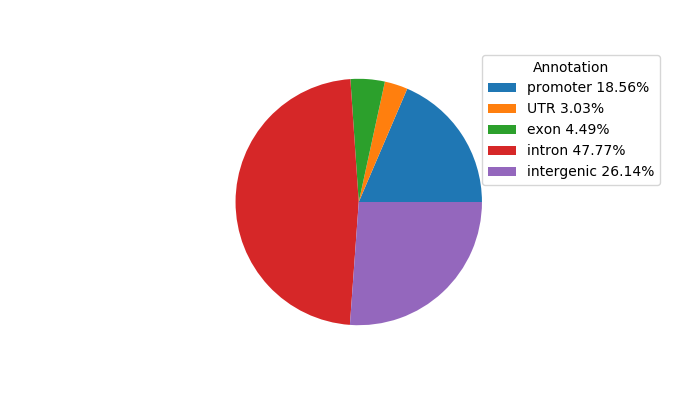
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_101882560_101882711 | 2.01 |
ZPLD1 |
zona pellucida-like domain containing 1 |
64547 |
0.14 |
| chr9_78830303_78830575 | 1.31 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
26884 |
0.26 |
| chrX_2815646_2815988 | 1.25 |
ARSD-AS1 |
ARSD antisense RNA 1 |
7128 |
0.17 |
| chr2_150023630_150023914 | 1.19 |
LYPD6B |
LY6/PLAUR domain containing 6B |
6782 |
0.33 |
| chr10_11629983_11630134 | 1.15 |
USP6NL |
USP6 N-terminal like |
14218 |
0.23 |
| chr8_28558220_28558688 | 1.15 |
EXTL3-AS1 |
EXTL3 antisense RNA 1 |
316 |
0.56 |
| chr3_128444618_128444816 | 1.14 |
RAB7A |
RAB7A, member RAS oncogene family |
248 |
0.92 |
| chr11_8742901_8743052 | 1.13 |
ST5 |
suppression of tumorigenicity 5 |
2286 |
0.2 |
| chr10_31321539_31321725 | 1.11 |
ZNF438 |
zinc finger protein 438 |
766 |
0.74 |
| chr1_209492900_209493051 | 1.08 |
ENSG00000230937 |
. |
112503 |
0.07 |
| chr12_48148703_48148929 | 1.06 |
SLC48A1 |
solute carrier family 48 (heme transporter), member 1 |
1117 |
0.4 |
| chr1_47180812_47180963 | 1.05 |
EFCAB14 |
EF-hand calcium binding domain 14 |
3849 |
0.2 |
| chr1_17572036_17572187 | 1.04 |
PADI3 |
peptidyl arginine deiminase, type III |
3482 |
0.2 |
| chr4_111106837_111106988 | 1.03 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
12897 |
0.25 |
| chr20_24387722_24387873 | 1.01 |
SYNDIG1 |
synapse differentiation inducing 1 |
62038 |
0.15 |
| chr8_81785077_81785279 | 1.00 |
ZNF704 |
zinc finger protein 704 |
722 |
0.78 |
| chr12_95071507_95071838 | 0.98 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
27334 |
0.23 |
| chr19_40596172_40596384 | 0.98 |
ZNF780A |
zinc finger protein 780A |
86 |
0.82 |
| chr2_34257_34408 | 0.97 |
FAM110C |
family with sequence similarity 110, member C |
12053 |
0.3 |
| chr6_86175199_86175350 | 0.97 |
NT5E |
5'-nucleotidase, ecto (CD73) |
5825 |
0.29 |
| chr1_159915900_159916142 | 0.95 |
IGSF9 |
immunoglobulin superfamily, member 9 |
635 |
0.57 |
| chr6_159224800_159225738 | 0.95 |
EZR-AS1 |
EZR antisense RNA 1 |
13774 |
0.17 |
| chr2_120041441_120041676 | 0.91 |
STEAP3-AS1 |
STEAP3 antisense RNA 1 |
34911 |
0.15 |
| chr11_126235331_126235482 | 0.90 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
9537 |
0.16 |
| chr5_55786776_55786927 | 0.90 |
CTC-236F12.4 |
Uncharacterized protein |
9255 |
0.21 |
| chr18_60499710_60499861 | 0.90 |
AC015989.2 |
|
6163 |
0.22 |
| chr11_110066760_110066911 | 0.88 |
RDX |
radixin |
41776 |
0.19 |
| chr17_34880270_34880421 | 0.88 |
MYO19 |
myosin XIX |
9701 |
0.12 |
| chr20_50104752_50104903 | 0.87 |
ENSG00000266761 |
. |
35313 |
0.19 |
| chr11_126252609_126252760 | 0.87 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
9386 |
0.18 |
| chr10_91217956_91218107 | 0.87 |
RP11-168O10.6 |
|
2210 |
0.28 |
| chr8_126239012_126239417 | 0.87 |
ENSG00000242170 |
. |
43632 |
0.17 |
| chr7_22473312_22473463 | 0.87 |
ENSG00000263927 |
. |
53764 |
0.13 |
| chr1_114888962_114889116 | 0.87 |
TRIM33 |
tripartite motif containing 33 |
116819 |
0.06 |
| chr14_61993184_61993433 | 0.87 |
RP11-47I22.4 |
|
2538 |
0.3 |
| chr6_18425072_18425223 | 0.86 |
ENSG00000238458 |
. |
22864 |
0.21 |
| chr10_119169187_119169338 | 0.85 |
PDZD8 |
PDZ domain containing 8 |
34284 |
0.17 |
| chr1_36040729_36040916 | 0.85 |
RP4-728D4.2 |
|
1474 |
0.29 |
| chr17_70412226_70412377 | 0.85 |
ENSG00000200783 |
. |
247990 |
0.02 |
| chr1_174935228_174935449 | 0.84 |
RABGAP1L |
RAB GTPase activating protein 1-like |
1433 |
0.35 |
| chr2_12695125_12695442 | 0.84 |
ENSG00000207183 |
. |
143656 |
0.05 |
| chr12_92790516_92790729 | 0.83 |
RP11-693J15.4 |
|
24685 |
0.17 |
| chr1_115781808_115781959 | 0.82 |
RP4-663N10.1 |
|
43772 |
0.18 |
| chr4_40911168_40911319 | 0.82 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
15954 |
0.2 |
| chr16_84390290_84390441 | 0.82 |
ATP2C2 |
ATPase, Ca++ transporting, type 2C, member 2 |
11768 |
0.18 |
| chr8_81665687_81665939 | 0.82 |
ENSG00000212429 |
. |
3885 |
0.28 |
| chr16_67960353_67960504 | 0.81 |
CTRL |
chymotrypsin-like |
5337 |
0.09 |
| chr6_167426333_167426484 | 0.81 |
FGFR1OP |
FGFR1 oncogene partner |
13512 |
0.14 |
| chr10_50626953_50627104 | 0.80 |
RP11-123B3.2 |
|
295 |
0.91 |
| chr6_108496924_108497159 | 0.80 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
7074 |
0.18 |
| chr21_43612405_43612556 | 0.80 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
7319 |
0.2 |
| chr3_193509513_193509757 | 0.80 |
ENSG00000243991 |
. |
51125 |
0.16 |
| chr17_79485610_79485838 | 0.80 |
RP13-766D20.2 |
|
342 |
0.76 |
| chr9_22104098_22104249 | 0.79 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
9504 |
0.26 |
| chr15_93592088_93592239 | 0.79 |
RGMA |
repulsive guidance molecule family member a |
24211 |
0.21 |
| chr14_59788218_59788466 | 0.79 |
DAAM1 |
dishevelled associated activator of morphogenesis 1 |
58169 |
0.14 |
| chr12_11883141_11883434 | 0.78 |
ETV6 |
ets variant 6 |
22148 |
0.26 |
| chr16_3012791_3012942 | 0.78 |
KREMEN2 |
kringle containing transmembrane protein 2 |
1079 |
0.26 |
| chr16_75130684_75130883 | 0.78 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
7899 |
0.16 |
| chr13_50932016_50932167 | 0.78 |
ENSG00000221198 |
. |
5282 |
0.33 |
| chr10_75815480_75815698 | 0.77 |
VCL |
vinculin |
27645 |
0.17 |
| chr4_101351551_101351881 | 0.77 |
EMCN |
endomucin |
44518 |
0.2 |
| chr16_85678818_85679245 | 0.76 |
GSE1 |
Gse1 coiled-coil protein |
9007 |
0.17 |
| chr9_71737791_71737961 | 0.76 |
TJP2 |
tight junction protein 2 |
1652 |
0.48 |
| chr19_54982368_54982633 | 0.75 |
CDC42EP5 |
CDC42 effector protein (Rho GTPase binding) 5 |
1911 |
0.17 |
| chr3_150461183_150461334 | 0.75 |
SIAH2-AS1 |
SIAH2 antisense RNA 1 |
18466 |
0.15 |
| chr11_95829326_95829477 | 0.75 |
MTMR2 |
myotubularin related protein 2 |
171942 |
0.03 |
| chr4_27031264_27031415 | 0.75 |
STIM2 |
stromal interaction molecule 2 |
21244 |
0.28 |
| chr11_71783035_71783766 | 0.75 |
ENSG00000263742 |
. |
52 |
0.94 |
| chr2_213642051_213642202 | 0.75 |
ENSG00000221388 |
. |
13727 |
0.3 |
| chr9_518186_518337 | 0.75 |
ENSG00000264949 |
. |
10708 |
0.14 |
| chr3_87367935_87368086 | 0.73 |
POU1F1 |
POU class 1 homeobox 1 |
42273 |
0.19 |
| chr17_19099525_19099830 | 0.73 |
ENSG00000264940 |
. |
6119 |
0.1 |
| chr1_202349043_202349194 | 0.73 |
ENSG00000272262 |
. |
30217 |
0.15 |
| chrX_7050343_7050602 | 0.73 |
ENSG00000264268 |
. |
15429 |
0.23 |
| chr8_102449532_102449683 | 0.72 |
ENSG00000239211 |
. |
39446 |
0.16 |
| chr18_52497228_52497379 | 0.72 |
RAB27B |
RAB27B, member RAS oncogene family |
1873 |
0.45 |
| chr7_2549187_2549338 | 0.72 |
LFNG |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
2901 |
0.22 |
| chr15_93365955_93366117 | 0.72 |
FAM174B |
family with sequence similarity 174, member B |
13008 |
0.16 |
| chr4_154388689_154389189 | 0.72 |
KIAA0922 |
KIAA0922 |
1438 |
0.51 |
| chr11_14375636_14375787 | 0.72 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
335 |
0.93 |
| chr13_50160162_50160384 | 0.71 |
RCBTB1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
554 |
0.8 |
| chr12_67355263_67355414 | 0.71 |
GRIP1 |
glutamate receptor interacting protein 1 |
157372 |
0.04 |
| chr17_4711269_4711435 | 0.71 |
PLD2 |
phospholipase D2 |
495 |
0.61 |
| chr17_35974821_35975108 | 0.71 |
RP11-697E22.1 |
|
5177 |
0.17 |
| chr7_116167296_116167447 | 0.70 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
1024 |
0.48 |
| chr2_191183153_191183456 | 0.70 |
HIBCH |
3-hydroxyisobutyryl-CoA hydrolase |
1312 |
0.49 |
| chr1_209816372_209816523 | 0.70 |
LAMB3 |
laminin, beta 3 |
8232 |
0.15 |
| chr17_79787756_79787907 | 0.70 |
FAM195B |
family with sequence similarity 195, member B |
3287 |
0.09 |
| chr1_25894144_25894295 | 0.70 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
24148 |
0.17 |
| chr2_61897669_61897960 | 0.69 |
ENSG00000206973 |
. |
65063 |
0.12 |
| chr21_16242942_16243093 | 0.69 |
AF127577.8 |
|
47838 |
0.16 |
| chr9_27634943_27635114 | 0.69 |
C9orf72 |
chromosome 9 open reading frame 72 |
61164 |
0.14 |
| chr9_6133801_6133990 | 0.69 |
IL33 |
interleukin 33 |
81910 |
0.09 |
| chr1_228280825_228280976 | 0.69 |
ENSG00000264944 |
. |
4064 |
0.14 |
| chr8_62008130_62008281 | 0.69 |
CLVS1 |
clavesin 1 |
38488 |
0.22 |
| chr19_12405667_12405818 | 0.69 |
ZNF44 |
zinc finger protein 44 |
40 |
0.97 |
| chr12_25921695_25921846 | 0.68 |
ENSG00000222950 |
. |
10733 |
0.28 |
| chr12_14537097_14537641 | 0.68 |
ATF7IP |
activating transcription factor 7 interacting protein |
629 |
0.79 |
| chr6_37103887_37104038 | 0.68 |
PIM1 |
pim-1 oncogene |
34017 |
0.15 |
| chr17_25833827_25833978 | 0.68 |
KSR1 |
kinase suppressor of ras 1 |
34866 |
0.16 |
| chr4_111283130_111283281 | 0.68 |
ENSG00000199894 |
. |
71139 |
0.11 |
| chr6_18053935_18054086 | 0.67 |
KIF13A |
kinesin family member 13A |
66156 |
0.11 |
| chr2_242209688_242209941 | 0.67 |
HDLBP |
high density lipoprotein binding protein |
1572 |
0.31 |
| chr16_81222783_81222934 | 0.67 |
PKD1L2 |
polycystic kidney disease 1-like 2 |
7924 |
0.19 |
| chr16_68268270_68268421 | 0.66 |
ESRP2 |
epithelial splicing regulatory protein 2 |
1690 |
0.16 |
| chr15_99206334_99206549 | 0.66 |
IGF1R |
insulin-like growth factor 1 receptor |
14168 |
0.22 |
| chr1_32135277_32135428 | 0.66 |
COL16A1 |
collagen, type XVI, alpha 1 |
3670 |
0.16 |
| chr10_127941521_127941672 | 0.65 |
ENSG00000222740 |
. |
107445 |
0.07 |
| chr8_37403860_37404011 | 0.65 |
RP11-150O12.6 |
|
29396 |
0.2 |
| chr21_46493345_46493835 | 0.65 |
AP001579.1 |
Uncharacterized protein |
663 |
0.49 |
| chr11_61463037_61463336 | 0.65 |
DAGLA |
diacylglycerol lipase, alpha |
15281 |
0.16 |
| chr10_99168304_99168455 | 0.65 |
RRP12 |
ribosomal RNA processing 12 homolog (S. cerevisiae) |
7252 |
0.1 |
| chr6_73839968_73840119 | 0.65 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
9763 |
0.19 |
| chr6_168436215_168436366 | 0.65 |
KIF25 |
kinesin family member 25 |
1612 |
0.45 |
| chr12_123187557_123187708 | 0.64 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
258 |
0.9 |
| chr4_170333365_170333516 | 0.64 |
SH3RF1 |
SH3 domain containing ring finger 1 |
141184 |
0.05 |
| chr1_115612637_115613031 | 0.64 |
TSPAN2 |
tetraspanin 2 |
19201 |
0.23 |
| chr4_177760307_177760458 | 0.64 |
VEGFC |
vascular endothelial growth factor C |
46501 |
0.2 |
| chr17_4542217_4542549 | 0.63 |
ALOX15 |
arachidonate 15-lipoxygenase |
794 |
0.48 |
| chr2_109236323_109236550 | 0.63 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
1286 |
0.53 |
| chr4_154004111_154004403 | 0.63 |
TRIM2 |
tripartite motif containing 2 |
69388 |
0.12 |
| chr1_68611188_68611339 | 0.63 |
GNG12-AS1 |
GNG12 antisense RNA 1 |
7223 |
0.23 |
| chr9_37084008_37084159 | 0.62 |
RP11-220I1.2 |
|
31404 |
0.15 |
| chr5_118703141_118703501 | 0.62 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
11591 |
0.21 |
| chr12_104194037_104194232 | 0.62 |
RP11-650K20.2 |
|
19254 |
0.13 |
| chr7_155526567_155526809 | 0.62 |
RBM33 |
RNA binding motif protein 33 |
12425 |
0.24 |
| chr8_56826165_56826541 | 0.61 |
ENSG00000216204 |
. |
4833 |
0.16 |
| chr11_119230682_119230833 | 0.61 |
USP2 |
ubiquitin specific peptidase 2 |
4160 |
0.1 |
| chr19_19706143_19706366 | 0.61 |
AC002306.1 |
|
10905 |
0.12 |
| chr17_74496103_74496254 | 0.61 |
RHBDF2 |
rhomboid 5 homolog 2 (Drosophila) |
1275 |
0.29 |
| chr17_19085056_19085346 | 0.60 |
ENSG00000263934 |
. |
6128 |
0.09 |
| chr19_6236985_6237136 | 0.60 |
CTC-503J8.4 |
|
22108 |
0.13 |
| chr3_67022512_67022756 | 0.60 |
KBTBD8 |
kelch repeat and BTB (POZ) domain containing 8 |
26093 |
0.28 |
| chr5_148926631_148926856 | 0.60 |
CSNK1A1 |
casein kinase 1, alpha 1 |
3119 |
0.21 |
| chr7_150450811_150450962 | 0.60 |
GIMAP5 |
GTPase, IMAP family member 5 |
16450 |
0.15 |
| chr5_11903259_11903410 | 0.59 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
20 |
0.99 |
| chr20_50177544_50177792 | 0.59 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
1500 |
0.54 |
| chr1_9144622_9144884 | 0.59 |
ENSG00000252404 |
. |
1998 |
0.27 |
| chr5_1793158_1793909 | 0.59 |
MRPL36 |
mitochondrial ribosomal protein L36 |
6331 |
0.19 |
| chr2_38920217_38920368 | 0.59 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
11855 |
0.17 |
| chr19_13273549_13273700 | 0.58 |
CTC-250I14.6 |
|
8617 |
0.11 |
| chr16_57300234_57300438 | 0.58 |
PLLP |
plasmolipin |
3053 |
0.17 |
| chr3_37326827_37326996 | 0.58 |
GOLGA4 |
golgin A4 |
3237 |
0.25 |
| chr14_34801203_34801354 | 0.58 |
EGLN3 |
egl-9 family hypoxia-inducible factor 3 |
109195 |
0.06 |
| chr2_31450999_31451150 | 0.58 |
ENSG00000201671 |
. |
104 |
0.97 |
| chr14_52332743_52333227 | 0.58 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
4943 |
0.23 |
| chr12_64798932_64799366 | 0.58 |
XPOT |
exportin, tRNA |
323 |
0.88 |
| chr18_39534789_39534940 | 0.58 |
PIK3C3 |
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
307 |
0.95 |
| chr10_106077859_106078132 | 0.58 |
RP11-127L20.5 |
|
6096 |
0.14 |
| chr6_76462249_76462400 | 0.58 |
MYO6 |
myosin VI |
3275 |
0.25 |
| chr2_102091009_102091179 | 0.57 |
RFX8 |
RFX family member 8, lacking RFX DNA binding domain |
71 |
0.98 |
| chr11_77113463_77113614 | 0.57 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
9411 |
0.24 |
| chr12_44201145_44201617 | 0.57 |
TWF1 |
twinfilin actin-binding protein 1 |
1203 |
0.49 |
| chr16_74483930_74484081 | 0.56 |
ENSG00000251794 |
. |
6434 |
0.16 |
| chr6_71109282_71109433 | 0.56 |
FAM135A |
family with sequence similarity 135, member A |
13654 |
0.21 |
| chr17_18973507_18973758 | 0.56 |
ENSG00000262074 |
. |
6183 |
0.11 |
| chr14_104583310_104583461 | 0.56 |
ENSG00000207568 |
. |
357 |
0.84 |
| chr8_22014494_22014879 | 0.56 |
LGI3 |
leucine-rich repeat LGI family, member 3 |
89 |
0.94 |
| chr15_62598892_62599117 | 0.56 |
RP11-299H22.5 |
|
36125 |
0.17 |
| chr11_70333823_70333974 | 0.56 |
AP001271.3 |
|
10201 |
0.17 |
| chr10_126410305_126410507 | 0.56 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
17653 |
0.16 |
| chr1_14087288_14087439 | 0.56 |
PRDM2 |
PR domain containing 2, with ZNF domain |
10858 |
0.25 |
| chr16_68670601_68670752 | 0.55 |
ENSG00000252640 |
. |
5805 |
0.13 |
| chr3_119807499_119807666 | 0.55 |
GSK3B |
glycogen synthase kinase 3 beta |
4931 |
0.22 |
| chr11_123325382_123325669 | 0.55 |
AP000783.1 |
Uncharacterized protein |
24467 |
0.2 |
| chrX_30918576_30918727 | 0.55 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
33332 |
0.17 |
| chr9_21546893_21547044 | 0.55 |
MIR31HG |
MIR31 host gene (non-protein coding) |
12700 |
0.16 |
| chr1_2347801_2347952 | 0.55 |
PEX10 |
peroxisomal biogenesis factor 10 |
2640 |
0.15 |
| chr1_100169867_100170018 | 0.55 |
PALMD |
palmdelphin |
58193 |
0.11 |
| chr8_103664408_103664722 | 0.55 |
KLF10 |
Kruppel-like factor 10 |
2038 |
0.38 |
| chr12_62671051_62671202 | 0.54 |
USP15 |
ubiquitin specific peptidase 15 |
1283 |
0.49 |
| chr6_6800951_6801164 | 0.54 |
ENSG00000240936 |
. |
138073 |
0.05 |
| chr2_181970504_181970655 | 0.54 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
123829 |
0.06 |
| chr3_185276204_185276355 | 0.54 |
LIPH |
lipase, member H |
5878 |
0.22 |
| chr12_120667953_120668326 | 0.54 |
PXN |
paxillin |
3492 |
0.15 |
| chr9_127030175_127030580 | 0.53 |
RP11-121A14.3 |
|
5222 |
0.19 |
| chr3_32861161_32861312 | 0.53 |
TRIM71 |
tripartite motif containing 71, E3 ubiquitin protein ligase |
1726 |
0.45 |
| chr9_116364960_116365111 | 0.53 |
RGS3 |
regulator of G-protein signaling 3 |
9269 |
0.22 |
| chr11_119038916_119039214 | 0.53 |
NLRX1 |
NLR family member X1 |
19 |
0.95 |
| chr2_192167664_192167815 | 0.53 |
MYO1B |
myosin IB |
26128 |
0.22 |
| chr10_102270443_102270627 | 0.53 |
SEC31B |
SEC31 homolog B (S. cerevisiae) |
9056 |
0.16 |
| chr6_50788742_50788893 | 0.53 |
TFAP2B |
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
2378 |
0.45 |
| chr6_110081297_110081448 | 0.52 |
FIG4 |
FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae) |
6560 |
0.28 |
| chr6_2542347_2542580 | 0.52 |
C6orf195 |
chromosome 6 open reading frame 195 |
92374 |
0.09 |
| chr22_20868530_20868681 | 0.52 |
MED15 |
mediator complex subunit 15 |
6261 |
0.11 |
| chr15_58896636_58896787 | 0.52 |
ENSG00000239100 |
. |
4277 |
0.22 |
| chr1_234862986_234863275 | 0.52 |
ENSG00000201638 |
. |
110590 |
0.06 |
| chr5_157340123_157340274 | 0.52 |
CLINT1 |
clathrin interactor 1 |
54015 |
0.13 |
| chr5_139702813_139703249 | 0.52 |
PFDN1 |
prefoldin subunit 1 |
20325 |
0.11 |
| chr1_178314482_178314633 | 0.52 |
RASAL2 |
RAS protein activator like 2 |
3951 |
0.34 |
| chr1_89727484_89727635 | 0.52 |
RP4-620F22.2 |
|
1294 |
0.45 |
| chr4_26344038_26344485 | 0.52 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
501 |
0.88 |
| chr9_132514460_132514833 | 0.52 |
PTGES |
prostaglandin E synthase |
680 |
0.65 |
| chr19_43959930_43960130 | 0.52 |
LYPD3 |
LY6/PLAUR domain containing 3 |
9782 |
0.14 |
| chr15_99401134_99401285 | 0.52 |
IGF1R |
insulin-like growth factor 1 receptor |
32361 |
0.17 |
| chr3_44690178_44690339 | 0.51 |
ZNF35 |
zinc finger protein 35 |
21 |
0.97 |
| chr18_54128502_54128653 | 0.51 |
TXNL1 |
thioredoxin-like 1 |
153145 |
0.04 |
| chr10_3929538_3929752 | 0.51 |
KLF6 |
Kruppel-like factor 6 |
102172 |
0.08 |
| chr9_130396397_130396548 | 0.51 |
STXBP1 |
syntaxin binding protein 1 |
21904 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.5 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.3 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.1 | 0.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.3 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.3 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.1 | 0.5 | GO:0043302 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.4 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.2 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.1 | 0.3 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.1 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.1 | 0.3 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 | 0.2 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.1 | 0.2 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.1 | 0.3 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.2 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.1 | 1.2 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.1 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.3 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.1 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.2 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0032048 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.2 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0055022 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.3 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.1 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.4 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0090195 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.2 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0051095 | regulation of helicase activity(GO:0051095) negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0031659 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 0.2 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.1 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.0 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:1903504 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.7 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.2 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.3 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.1 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.0 | GO:0002248 | wound healing involved in inflammatory response(GO:0002246) connective tissue replacement involved in inflammatory response wound healing(GO:0002248) inflammatory response to wounding(GO:0090594) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0071333 | cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.5 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0098868 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.0 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.0 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.1 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.1 | GO:2001234 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.3 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.0 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0046036 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0046885 | regulation of hormone biosynthetic process(GO:0046885) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.0 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.1 | GO:0046348 | amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.0 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 | 0.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.1 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0006693 | prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0044091 | membrane biogenesis(GO:0044091) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.0 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.1 | GO:0032372 | negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.1 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.0 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.0 | GO:0060192 | negative regulation of lipase activity(GO:0060192) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.3 | GO:0043928 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.3 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.3 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.1 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 0.2 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.1 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0035121 | obsolete tail morphogenesis(GO:0035121) |
| 0.0 | 0.3 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.0 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.0 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.4 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.0 | GO:0048867 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:0032232 | negative regulation of actin filament bundle assembly(GO:0032232) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 1.9 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.3 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.1 | 0.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.2 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.5 | GO:0052742 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol kinase activity(GO:0052742) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0046527 | UDP-glucosyltransferase activity(GO:0035251) glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.3 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) poly-purine tract binding(GO:0070717) |
| 0.0 | 0.0 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.6 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0047115 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |