Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
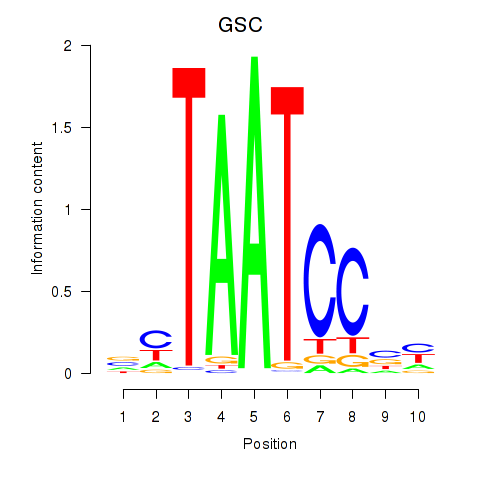
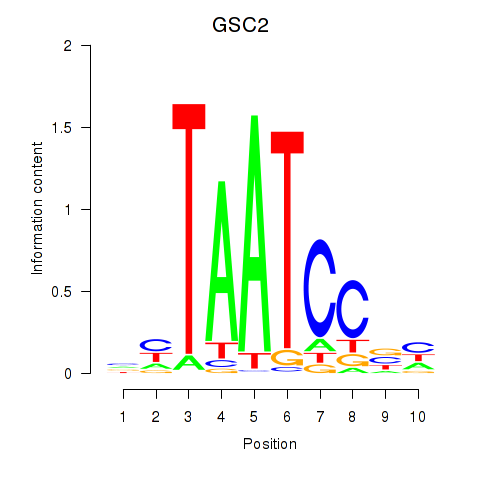
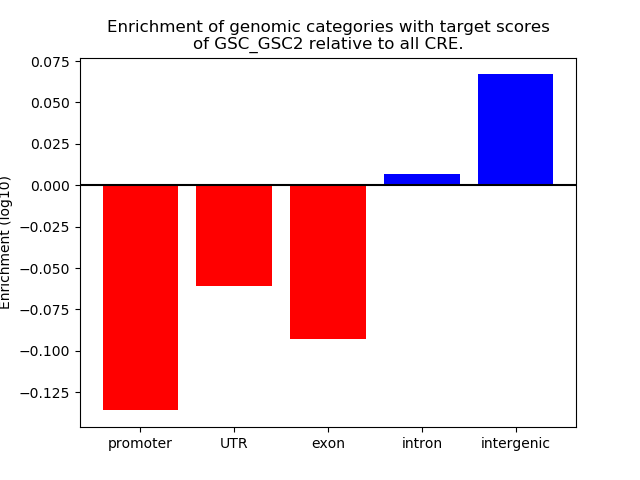
Results for GSC_GSC2
Z-value: 1.33
Transcription factors associated with GSC_GSC2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GSC
|
ENSG00000133937.3 | goosecoid homeobox |
|
GSC2
|
ENSG00000063515.2 | goosecoid homeobox 2 |
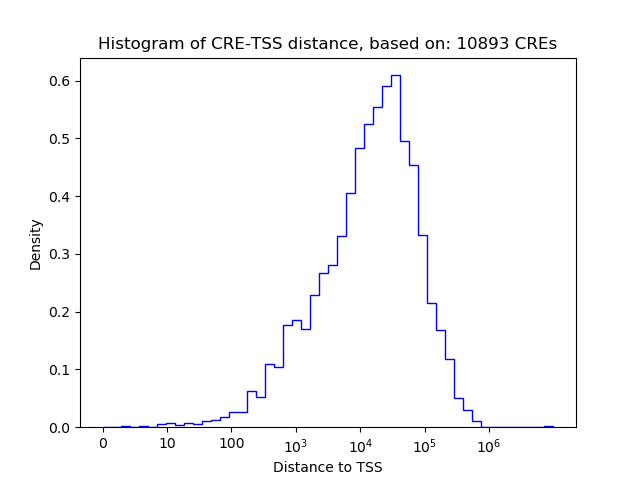
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_95205498_95205649 | GSC | 30989 | 0.202484 | 0.81 | 8.6e-03 | Click! |
| chr14_95236567_95236718 | GSC | 80 | 0.983162 | -0.71 | 3.1e-02 | Click! |
| chr14_95237974_95238125 | GSC | 1487 | 0.529948 | -0.57 | 1.1e-01 | Click! |
| chr14_95205255_95205406 | GSC | 31232 | 0.201656 | 0.55 | 1.2e-01 | Click! |
| chr14_95235460_95235620 | GSC | 1022 | 0.660084 | 0.49 | 1.8e-01 | Click! |
| chr22_19136647_19137197 | GSC2 | 874 | 0.433300 | -0.70 | 3.5e-02 | Click! |
| chr22_19142720_19142871 | GSC2 | 4999 | 0.121362 | -0.57 | 1.1e-01 | Click! |
| chr22_19136477_19136628 | GSC2 | 1244 | 0.304480 | -0.45 | 2.2e-01 | Click! |
| chr22_19136086_19136237 | GSC2 | 1635 | 0.231016 | -0.23 | 5.6e-01 | Click! |
| chr22_19137289_19137440 | GSC2 | 432 | 0.720068 | -0.09 | 8.2e-01 | Click! |
Activity of the GSC_GSC2 motif across conditions
Conditions sorted by the z-value of the GSC_GSC2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
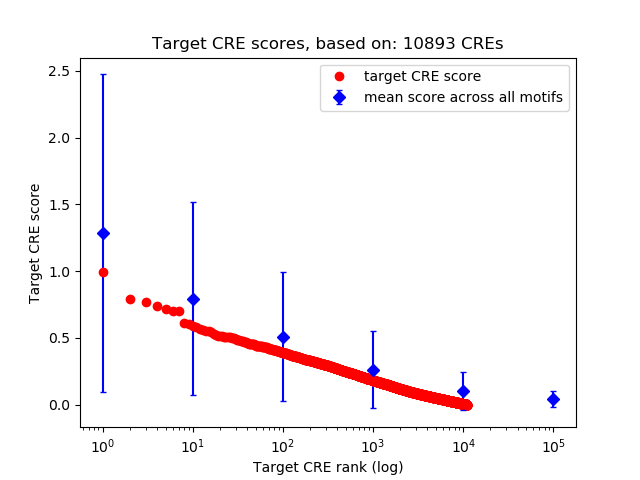
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_201426596_201426747 | 0.99 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
11641 |
0.15 |
| chr12_11733022_11733284 | 0.79 |
ENSG00000251747 |
. |
33740 |
0.16 |
| chr2_43152776_43153425 | 0.77 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
133368 |
0.05 |
| chr5_148352238_148352777 | 0.74 |
RP11-44B19.1 |
|
2604 |
0.3 |
| chr14_69544397_69544681 | 0.71 |
ENSG00000206768 |
. |
66131 |
0.1 |
| chr3_170470237_170470470 | 0.70 |
ENSG00000222411 |
. |
89608 |
0.08 |
| chr14_53301825_53302222 | 0.70 |
FERMT2 |
fermitin family member 2 |
29216 |
0.15 |
| chr3_174156335_174156660 | 0.62 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
2280 |
0.47 |
| chr9_127162086_127162596 | 0.60 |
PSMB7 |
proteasome (prosome, macropain) subunit, beta type, 7 |
15377 |
0.19 |
| chr2_45926277_45926449 | 0.59 |
U51244.2 |
|
23748 |
0.22 |
| chr5_159305915_159306327 | 0.58 |
ADRA1B |
adrenoceptor alpha 1B |
37669 |
0.2 |
| chr4_171593437_171593770 | 0.57 |
ENSG00000251961 |
. |
219015 |
0.02 |
| chr2_162949372_162949523 | 0.56 |
DPP4 |
dipeptidyl-peptidase 4 |
18395 |
0.18 |
| chr8_97603049_97603490 | 0.55 |
SDC2 |
syndecan 2 |
4654 |
0.31 |
| chr9_14271441_14271727 | 0.55 |
NFIB |
nuclear factor I/B |
36428 |
0.2 |
| chr1_59481843_59482033 | 0.54 |
JUN |
jun proto-oncogene |
232153 |
0.02 |
| chr10_31939351_31939502 | 0.53 |
ENSG00000222412 |
. |
105638 |
0.07 |
| chr1_52024489_52024927 | 0.53 |
OSBPL9 |
oxysterol binding protein-like 9 |
18143 |
0.15 |
| chr11_111428191_111428342 | 0.52 |
LAYN |
layilin |
15989 |
0.12 |
| chr1_30185013_30185164 | 0.52 |
ENSG00000221126 |
. |
67563 |
0.14 |
| chr17_73690444_73690713 | 0.51 |
SAP30BP |
SAP30 binding protein |
1952 |
0.18 |
| chr2_159996739_159997187 | 0.51 |
ENSG00000202029 |
. |
113309 |
0.06 |
| chr1_85805501_85805652 | 0.51 |
ENSG00000264380 |
. |
55283 |
0.09 |
| chr15_46162513_46162674 | 0.51 |
SQRDL |
sulfide quinone reductase-like (yeast) |
187855 |
0.03 |
| chr1_21478340_21478712 | 0.51 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
22704 |
0.19 |
| chr9_117715769_117716310 | 0.51 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
23342 |
0.24 |
| chr2_174889480_174890119 | 0.50 |
SP3 |
Sp3 transcription factor |
59369 |
0.15 |
| chr17_30068402_30068553 | 0.50 |
ENSG00000202026 |
. |
27705 |
0.19 |
| chr6_3517760_3517987 | 0.50 |
SLC22A23 |
solute carrier family 22, member 23 |
60617 |
0.14 |
| chr9_132096828_132097338 | 0.49 |
ENSG00000242281 |
. |
35657 |
0.16 |
| chr17_40481304_40481455 | 0.49 |
STAT5A |
signal transducer and activator of transcription 5A |
23191 |
0.11 |
| chr7_47366010_47366161 | 0.49 |
TNS3 |
tensin 3 |
46169 |
0.21 |
| chr5_64493902_64494516 | 0.48 |
ENSG00000207439 |
. |
75013 |
0.12 |
| chr15_38288177_38288328 | 0.48 |
TMCO5A |
transmembrane and coiled-coil domains 5A |
60794 |
0.16 |
| chr9_114727126_114727277 | 0.48 |
ENSG00000222356 |
. |
31818 |
0.15 |
| chr5_39093457_39093608 | 0.48 |
AC008964.1 |
|
11826 |
0.23 |
| chr9_16143627_16143778 | 0.47 |
C9orf92 |
chromosome 9 open reading frame 92 |
72195 |
0.13 |
| chr4_152666058_152666209 | 0.47 |
PET112 |
PET112 homolog (yeast) |
16004 |
0.24 |
| chr2_43496469_43496754 | 0.47 |
AC010883.5 |
|
39899 |
0.17 |
| chr3_120147916_120148067 | 0.46 |
FSTL1 |
follistatin-like 1 |
21847 |
0.22 |
| chr8_37783052_37783203 | 0.46 |
GOT1L1 |
glutamic-oxaloacetic transaminase 1-like 1 |
11204 |
0.13 |
| chr5_149433185_149433336 | 0.46 |
ENSG00000238369 |
. |
24245 |
0.12 |
| chr3_69813399_69813761 | 0.46 |
MITF |
microphthalmia-associated transcription factor |
618 |
0.78 |
| chr4_77468211_77468426 | 0.46 |
ENSG00000207307 |
. |
14838 |
0.16 |
| chr22_22109341_22109765 | 0.46 |
YPEL1 |
yippee-like 1 (Drosophila) |
19430 |
0.12 |
| chr18_30500917_30501068 | 0.46 |
RP11-680N20.1 |
|
33047 |
0.21 |
| chr5_64500241_64500433 | 0.45 |
ENSG00000207439 |
. |
81141 |
0.11 |
| chr9_73518177_73518328 | 0.45 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
34278 |
0.22 |
| chr10_3570543_3570694 | 0.45 |
RP11-184A2.3 |
|
222641 |
0.02 |
| chr19_47478570_47478952 | 0.44 |
ENSG00000252071 |
. |
27215 |
0.14 |
| chr16_30724842_30725097 | 0.44 |
ENSG00000206755 |
. |
3111 |
0.11 |
| chr12_15785962_15786113 | 0.44 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
29171 |
0.19 |
| chr9_16795794_16795945 | 0.44 |
BNC2 |
basonuclin 2 |
36417 |
0.21 |
| chr10_35021138_35021390 | 0.44 |
PARD3 |
par-3 family cell polarity regulator |
82985 |
0.1 |
| chr8_119017458_119017609 | 0.44 |
EXT1 |
exostosin glycosyltransferase 1 |
105120 |
0.08 |
| chr6_43088179_43088500 | 0.44 |
PTK7 |
protein tyrosine kinase 7 |
9990 |
0.12 |
| chr14_55242379_55242669 | 0.44 |
SAMD4A |
sterile alpha motif domain containing 4A |
20973 |
0.23 |
| chr11_131928229_131928481 | 0.44 |
RP11-697E14.2 |
|
73800 |
0.12 |
| chr3_170820342_170820493 | 0.44 |
ENSG00000207963 |
. |
4131 |
0.3 |
| chr5_156943457_156943608 | 0.44 |
ADAM19 |
ADAM metallopeptidase domain 19 |
13702 |
0.17 |
| chr10_80886709_80887280 | 0.43 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
58202 |
0.14 |
| chr2_44357819_44358004 | 0.43 |
ENSG00000252599 |
. |
24123 |
0.17 |
| chr9_111396759_111396910 | 0.43 |
ACTL7B |
actin-like 7B |
222405 |
0.02 |
| chrX_12378398_12378549 | 0.43 |
FRMPD4-AS1 |
FRMPD4 antisense RNA 1 |
14779 |
0.29 |
| chr5_98396799_98396950 | 0.43 |
ENSG00000200351 |
. |
124423 |
0.06 |
| chr15_69850349_69850500 | 0.43 |
ENSG00000207119 |
. |
99936 |
0.06 |
| chr16_11098719_11098870 | 0.43 |
CLEC16A |
C-type lectin domain family 16, member A |
60356 |
0.08 |
| chr12_68141751_68141902 | 0.43 |
RP11-335O4.3 |
|
77127 |
0.11 |
| chr11_108786966_108787117 | 0.42 |
ENSG00000201243 |
. |
89203 |
0.1 |
| chr12_66341786_66341937 | 0.42 |
RP11-366L20.3 |
|
398 |
0.86 |
| chr5_34487773_34488055 | 0.42 |
RAI14 |
retinoic acid induced 14 |
168428 |
0.03 |
| chr20_43988664_43988912 | 0.42 |
SYS1 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
1789 |
0.23 |
| chr3_159645626_159645969 | 0.42 |
IL12A |
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
60740 |
0.1 |
| chr2_226928659_226928810 | 0.42 |
ENSG00000263363 |
. |
594775 |
0.0 |
| chr11_130752561_130752712 | 0.42 |
SNX19 |
sorting nexin 19 |
11133 |
0.3 |
| chr6_52376418_52376603 | 0.41 |
TRAM2 |
translocation associated membrane protein 2 |
65203 |
0.11 |
| chr7_55089353_55089504 | 0.41 |
EGFR |
epidermal growth factor receptor |
2617 |
0.41 |
| chr2_113470510_113470661 | 0.41 |
NT5DC4 |
5'-nucleotidase domain containing 4 |
8478 |
0.2 |
| chr11_111077610_111077761 | 0.41 |
C11orf53 |
chromosome 11 open reading frame 53 |
49022 |
0.14 |
| chr5_142286909_142287060 | 0.41 |
ARHGAP26 |
Rho GTPase activating protein 26 |
97 |
0.98 |
| chr6_138929947_138930098 | 0.41 |
NHSL1 |
NHS-like 1 |
36345 |
0.19 |
| chr17_2082426_2082658 | 0.41 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
6070 |
0.13 |
| chr7_100422531_100422682 | 0.41 |
EPHB4 |
EPH receptor B4 |
2131 |
0.18 |
| chr4_122712483_122712634 | 0.41 |
EXOSC9 |
exosome component 9 |
9914 |
0.16 |
| chr20_1809000_1809151 | 0.41 |
SIRPA |
signal-regulatory protein alpha |
66079 |
0.11 |
| chr1_157983762_157983913 | 0.40 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
11503 |
0.21 |
| chr5_149886580_149886731 | 0.40 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
1019 |
0.53 |
| chr20_33292677_33293239 | 0.40 |
TP53INP2 |
tumor protein p53 inducible nuclear protein 2 |
419 |
0.86 |
| chr6_1575312_1575463 | 0.40 |
FOXC1 |
forkhead box C1 |
35294 |
0.21 |
| chr20_7821639_7821790 | 0.40 |
HAO1 |
hydroxyacid oxidase (glycolate oxidase) 1 |
99407 |
0.08 |
| chr1_26797459_26798053 | 0.40 |
HMGN2 |
high mobility group nucleosomal binding domain 2 |
1254 |
0.35 |
| chr6_82041474_82041625 | 0.40 |
RP1-300G12.2 |
|
203081 |
0.03 |
| chr3_98613485_98613636 | 0.39 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
6455 |
0.19 |
| chr9_79316323_79316721 | 0.39 |
PRUNE2 |
prune homolog 2 (Drosophila) |
8631 |
0.24 |
| chr1_37937094_37937368 | 0.39 |
LINC01137 |
long intergenic non-protein coding RNA 1137 |
2781 |
0.2 |
| chr5_143022321_143022472 | 0.39 |
ENSG00000266478 |
. |
37029 |
0.18 |
| chr7_40619322_40619515 | 0.39 |
AC004988.1 |
|
32891 |
0.25 |
| chr2_220554157_220554308 | 0.39 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
58380 |
0.08 |
| chr2_217180660_217180876 | 0.39 |
AC069155.1 |
|
12720 |
0.18 |
| chr7_13920733_13920884 | 0.39 |
ETV1 |
ets variant 1 |
105258 |
0.08 |
| chr2_9391597_9391776 | 0.39 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
44792 |
0.18 |
| chr1_46650137_46650288 | 0.39 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
8052 |
0.12 |
| chr1_211210156_211210307 | 0.39 |
KCNH1-IT1 |
KCNH1 intronic transcript 1 (non-protein coding) |
96485 |
0.08 |
| chr9_37943494_37943645 | 0.39 |
ENSG00000251745 |
. |
6804 |
0.24 |
| chr1_94429653_94429804 | 0.39 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
47453 |
0.13 |
| chr10_104387429_104387633 | 0.39 |
TRIM8 |
tripartite motif containing 8 |
16722 |
0.16 |
| chr11_130642757_130642947 | 0.39 |
C11orf44 |
chromosome 11 open reading frame 44 |
100001 |
0.08 |
| chr17_40568875_40569026 | 0.39 |
PTRF |
polymerase I and transcript release factor |
6585 |
0.11 |
| chr14_103889997_103890148 | 0.39 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
37496 |
0.12 |
| chr6_147234985_147235152 | 0.38 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
1953 |
0.49 |
| chr3_52205760_52205911 | 0.38 |
POC1A |
POC1 centriolar protein A |
17129 |
0.12 |
| chr6_35393359_35394053 | 0.38 |
FANCE |
Fanconi anemia, complementation group E |
26432 |
0.15 |
| chr8_61863719_61864009 | 0.38 |
CLVS1 |
clavesin 1 |
105853 |
0.07 |
| chr10_131821141_131821577 | 0.38 |
EBF3 |
early B-cell factor 3 |
59254 |
0.15 |
| chr7_151572690_151573025 | 0.38 |
PRKAG2-AS1 |
PRKAG2 antisense RNA 1 |
1270 |
0.31 |
| chr5_159969265_159969416 | 0.38 |
ENSG00000253522 |
. |
56981 |
0.1 |
| chr8_119778762_119778913 | 0.38 |
SAMD12 |
sterile alpha motif domain containing 12 |
144603 |
0.05 |
| chr1_208396700_208396908 | 0.38 |
PLXNA2 |
plexin A2 |
20861 |
0.29 |
| chr11_78511494_78511645 | 0.38 |
TENM4 |
teneurin transmembrane protein 4 |
4887 |
0.29 |
| chr2_228741295_228741446 | 0.37 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
4991 |
0.23 |
| chr16_68333106_68333257 | 0.37 |
SLC7A6OS |
solute carrier family 7, member 6 opposite strand |
4951 |
0.09 |
| chr1_208382716_208382867 | 0.37 |
PLXNA2 |
plexin A2 |
34874 |
0.24 |
| chr3_149954251_149954402 | 0.37 |
RP11-167H9.4 |
|
138507 |
0.04 |
| chr17_43205364_43205515 | 0.37 |
PLCD3 |
phospholipase C, delta 3 |
4452 |
0.12 |
| chr17_19651678_19651829 | 0.37 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
7 |
0.97 |
| chr5_149887543_149887842 | 0.37 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
18 |
0.98 |
| chr17_79532915_79533078 | 0.37 |
NPLOC4 |
nuclear protein localization 4 homolog (S. cerevisiae) |
632 |
0.54 |
| chr1_9951525_9951676 | 0.37 |
CTNNBIP1 |
catenin, beta interacting protein 1 |
1695 |
0.29 |
| chr8_19045282_19045433 | 0.37 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
103117 |
0.08 |
| chr11_63750040_63750285 | 0.37 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
3307 |
0.14 |
| chr4_187606105_187606371 | 0.37 |
FAT1 |
FAT atypical cadherin 1 |
38771 |
0.2 |
| chr8_123772200_123772351 | 0.37 |
ZHX2 |
zinc fingers and homeoboxes 2 |
21358 |
0.21 |
| chr1_234700121_234700537 | 0.36 |
ENSG00000212144 |
. |
28692 |
0.16 |
| chr1_152022964_152023546 | 0.36 |
S100A11 |
S100 calcium binding protein A11 |
13744 |
0.14 |
| chr15_41573424_41573575 | 0.36 |
CHP1 |
calcineurin-like EF-hand protein 1 |
24391 |
0.13 |
| chr3_124277818_124277969 | 0.36 |
KALRN |
kalirin, RhoGEF kinase |
25613 |
0.23 |
| chr11_86447090_86447241 | 0.36 |
PRSS23 |
protease, serine, 23 |
54936 |
0.15 |
| chr16_66652366_66652517 | 0.36 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
13017 |
0.11 |
| chr11_108908571_108908861 | 0.36 |
ENSG00000201243 |
. |
32472 |
0.23 |
| chr6_22367405_22367556 | 0.36 |
PRL |
prolactin |
69750 |
0.13 |
| chr5_10478976_10479127 | 0.36 |
RP11-1C1.7 |
|
432 |
0.81 |
| chr1_225959781_225959947 | 0.36 |
SRP9 |
signal recognition particle 9kDa |
5667 |
0.16 |
| chr22_27602198_27602349 | 0.36 |
ENSG00000200443 |
. |
170423 |
0.04 |
| chr8_91532720_91532871 | 0.36 |
ENSG00000199354 |
. |
9439 |
0.26 |
| chr1_214731522_214731884 | 0.36 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
7137 |
0.28 |
| chr17_1690708_1690859 | 0.36 |
SMYD4 |
SET and MYND domain containing 4 |
13316 |
0.11 |
| chr2_28497749_28498516 | 0.36 |
AC093690.1 |
|
35194 |
0.19 |
| chr3_149469081_149469660 | 0.36 |
COMMD2 |
COMM domain containing 2 |
916 |
0.55 |
| chr3_185450728_185450956 | 0.36 |
C3orf65 |
chromosome 3 open reading frame 65 |
19762 |
0.2 |
| chr12_77425923_77426193 | 0.35 |
E2F7 |
E2F transcription factor 7 |
18468 |
0.27 |
| chr3_123979690_123979841 | 0.35 |
KALRN |
kalirin, RhoGEF kinase |
32928 |
0.22 |
| chr8_129187891_129188042 | 0.35 |
ENSG00000221261 |
. |
25604 |
0.21 |
| chr15_83611739_83611890 | 0.35 |
HOMER2 |
homer homolog 2 (Drosophila) |
9659 |
0.14 |
| chr1_205114875_205115026 | 0.35 |
RBBP5 |
retinoblastoma binding protein 5 |
23807 |
0.16 |
| chr10_91043296_91043536 | 0.35 |
RP11-149I23.3 |
|
106 |
0.95 |
| chr5_131600580_131600731 | 0.35 |
PDLIM4 |
PDZ and LIM domain 4 |
3513 |
0.2 |
| chr6_149199848_149199999 | 0.35 |
RP11-162J8.2 |
|
85897 |
0.09 |
| chr10_79709340_79709491 | 0.35 |
DLG5-AS1 |
DLG5 antisense RNA 1 |
22285 |
0.16 |
| chr6_39855140_39855437 | 0.35 |
RP11-61I13.3 |
|
944 |
0.63 |
| chr12_110388287_110388838 | 0.35 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
45467 |
0.11 |
| chr5_139069097_139069307 | 0.35 |
CXXC5 |
CXXC finger protein 5 |
9934 |
0.2 |
| chr5_77902045_77902196 | 0.35 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
42528 |
0.21 |
| chr5_180213979_180214228 | 0.35 |
CTC-205M6.5 |
|
766 |
0.61 |
| chr16_54025361_54025773 | 0.35 |
RP11-357N13.2 |
|
8314 |
0.19 |
| chr12_53465095_53465401 | 0.35 |
SPRYD3 |
SPRY domain containing 3 |
7913 |
0.11 |
| chr10_30236708_30236859 | 0.34 |
KIAA1462 |
KIAA1462 |
111670 |
0.07 |
| chr15_74629333_74629484 | 0.34 |
CCDC33 |
coiled-coil domain containing 33 |
18497 |
0.13 |
| chr6_155703153_155703304 | 0.34 |
TFB1M |
transcription factor B1, mitochondrial |
67610 |
0.11 |
| chr8_12583298_12583449 | 0.34 |
ENSG00000266206 |
. |
1440 |
0.38 |
| chr13_111411891_111412042 | 0.34 |
ING1 |
inhibitor of growth family, member 1 |
44637 |
0.15 |
| chr15_32993894_32994045 | 0.34 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
16206 |
0.17 |
| chr5_82772375_82772526 | 0.34 |
VCAN |
versican |
4706 |
0.32 |
| chr6_3743005_3743681 | 0.34 |
PXDC1 |
PX domain containing 1 |
4215 |
0.25 |
| chr19_17218227_17218378 | 0.34 |
MYO9B |
myosin IXB |
5774 |
0.13 |
| chr5_111004543_111004694 | 0.34 |
ENSG00000253057 |
. |
51140 |
0.14 |
| chr11_12307449_12307766 | 0.34 |
MICALCL |
MICAL C-terminal like |
840 |
0.61 |
| chr4_24227998_24228149 | 0.34 |
ENSG00000222262 |
. |
194851 |
0.03 |
| chr1_200341478_200341663 | 0.34 |
ZNF281 |
zinc finger protein 281 |
37550 |
0.22 |
| chr9_14251628_14251779 | 0.34 |
NFIB |
nuclear factor I/B |
56309 |
0.15 |
| chr2_227525621_227525772 | 0.34 |
ENSG00000263363 |
. |
2187 |
0.45 |
| chr5_395973_396219 | 0.34 |
AHRR |
aryl-hydrocarbon receptor repressor |
24939 |
0.14 |
| chr13_101959109_101959260 | 0.34 |
NALCN |
sodium leak channel, non-selective |
92321 |
0.09 |
| chr6_14286162_14286313 | 0.33 |
ENSG00000238987 |
. |
129544 |
0.06 |
| chr12_53693833_53694662 | 0.33 |
RP11-680A11.5 |
|
149 |
0.73 |
| chr3_53277561_53277865 | 0.33 |
TKT |
transketolase |
12303 |
0.14 |
| chr2_169371779_169371930 | 0.33 |
CERS6 |
ceramide synthase 6 |
59095 |
0.11 |
| chr21_37575111_37575262 | 0.33 |
ENSG00000265882 |
. |
10950 |
0.15 |
| chr18_46148945_46149096 | 0.33 |
ENSG00000266276 |
. |
47951 |
0.15 |
| chr8_101471248_101471399 | 0.33 |
KB-1615E4.2 |
|
16580 |
0.18 |
| chr15_40384302_40384677 | 0.33 |
RP11-521C20.3 |
|
3456 |
0.2 |
| chr12_52182216_52182367 | 0.33 |
AC068987.1 |
HCG1997999; cDNA FLJ33996 fis, clone DFNES2008881 |
21498 |
0.2 |
| chr4_77956814_77956965 | 0.33 |
SEPT11 |
septin 11 |
7019 |
0.23 |
| chr11_130652591_130652742 | 0.33 |
C11orf44 |
chromosome 11 open reading frame 44 |
109815 |
0.07 |
| chr1_109934550_109935130 | 0.33 |
SORT1 |
sortilin 1 |
1139 |
0.45 |
| chr10_79150233_79150384 | 0.33 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
13434 |
0.23 |
| chr1_154379358_154380147 | 0.33 |
RP11-350G8.5 |
|
712 |
0.52 |
| chr14_69522934_69523085 | 0.33 |
ACTN1-AS1 |
ACTN1 antisense RNA 1 |
76251 |
0.09 |
| chr6_113330842_113330993 | 0.33 |
ENSG00000201386 |
. |
38222 |
0.23 |
| chr1_223584357_223584508 | 0.33 |
C1orf65 |
chromosome 1 open reading frame 65 |
17717 |
0.2 |
| chr1_150481830_150481981 | 0.33 |
ECM1 |
extracellular matrix protein 1 |
1322 |
0.29 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.3 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.3 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.1 | 0.3 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.2 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.1 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.3 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 0.2 | GO:0019348 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.1 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.3 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.2 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.2 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.1 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.1 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.2 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 0.2 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.1 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0045628 | regulation of T-helper 2 cell differentiation(GO:0045628) |
| 0.0 | 0.2 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.3 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0048342 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.2 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.2 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.1 | GO:2000095 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0017000 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.1 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.0 | 0.1 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.2 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0090030 | regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.0 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0032048 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.0 | GO:2000647 | negative regulation of stem cell proliferation(GO:2000647) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.0 | GO:0009151 | purine deoxyribonucleotide metabolic process(GO:0009151) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.2 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.1 | GO:0046049 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.0 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 0.0 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.0 | 0.1 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.2 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.1 | GO:0046398 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.0 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.2 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.0 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.3 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0072666 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.0 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.0 | GO:0072193 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.3 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0055094 | response to lipoprotein particle(GO:0055094) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0034227 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.7 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.0 | GO:0090493 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.0 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.1 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0060841 | venous blood vessel morphogenesis(GO:0048845) venous blood vessel development(GO:0060841) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:0031427 | response to methotrexate(GO:0031427) |
| 0.0 | 0.1 | GO:0001963 | synaptic transmission, dopaminergic(GO:0001963) |
| 0.0 | 0.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.2 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0042597 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 1.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.0 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.3 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.1 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.2 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.2 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0032356 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.0 | REACTOME IMMUNE SYSTEM | Genes involved in Immune System |
| 0.0 | 0.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | Genes involved in Negative regulation of FGFR signaling |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.0 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |