Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for GTF2I
Z-value: 1.20
Transcription factors associated with GTF2I
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
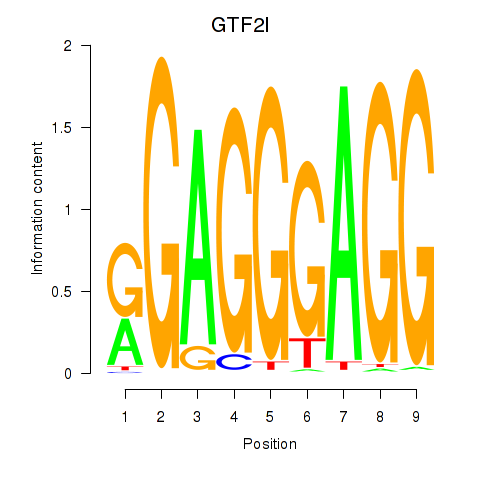
GTF2I
|
ENSG00000077809.8 | general transcription factor IIi |
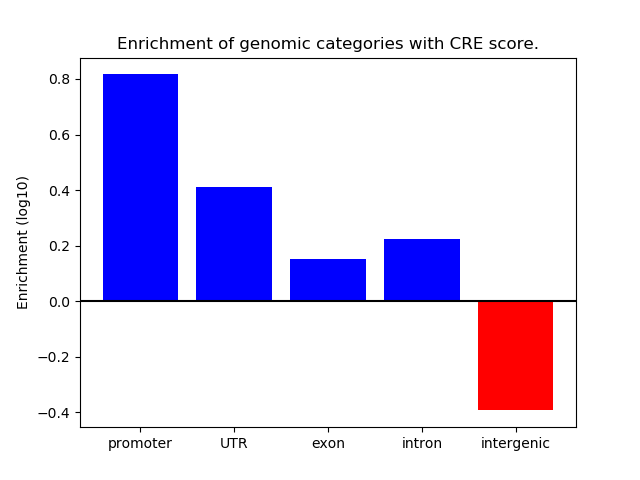
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_74071229_74071632 | GTF2I | 581 | 0.795860 | -0.80 | 1.0e-02 | Click! |
| chr7_74081638_74081789 | GTF2I | 9356 | 0.225626 | -0.80 | 1.0e-02 | Click! |
| chr7_74048042_74048193 | GTF2I | 23894 | 0.190203 | -0.77 | 1.4e-02 | Click! |
| chr7_74074150_74074543 | GTF2I | 1989 | 0.380716 | -0.74 | 2.2e-02 | Click! |
| chr7_74057428_74057605 | GTF2I | 14495 | 0.214789 | -0.73 | 2.7e-02 | Click! |
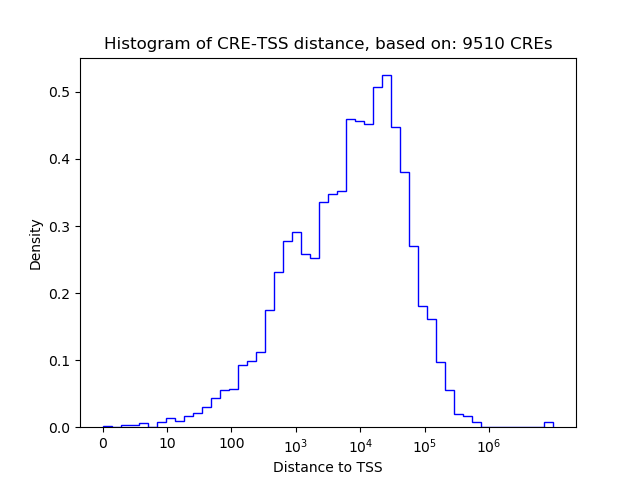
Activity of the GTF2I motif across conditions
Conditions sorted by the z-value of the GTF2I motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
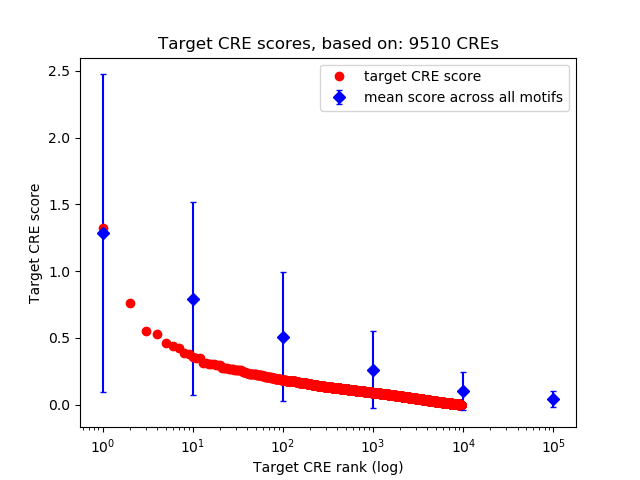
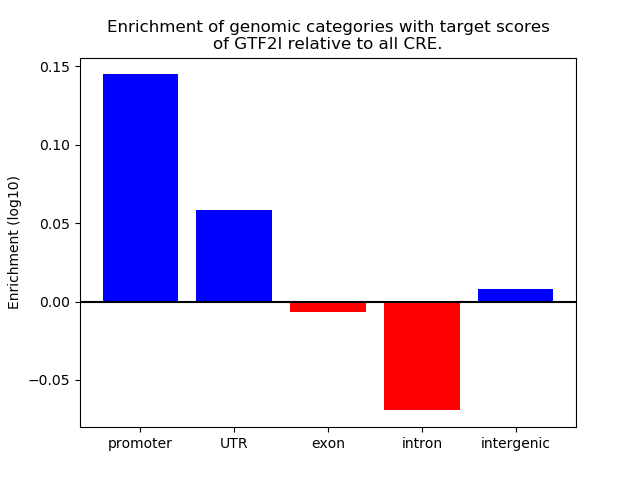
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_10417901_10418493 | 1.33 |
ENSG00000264030 |
. |
2560 |
0.25 |
| chr19_7989733_7990535 | 0.76 |
CTD-3193O13.8 |
|
752 |
0.29 |
| chr8_140716343_140716851 | 0.55 |
KCNK9 |
potassium channel, subfamily K, member 9 |
1298 |
0.63 |
| chr9_94579747_94580327 | 0.53 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
131125 |
0.05 |
| chr7_2672947_2673687 | 0.46 |
TTYH3 |
tweety family member 3 |
1527 |
0.39 |
| chr21_47714776_47715544 | 0.44 |
YBEY |
ybeY metallopeptidase (putative) |
8601 |
0.11 |
| chr14_103986978_103987655 | 0.42 |
CKB |
creatine kinase, brain |
122 |
0.92 |
| chr3_13045853_13046441 | 0.39 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
17611 |
0.25 |
| chr17_38500621_38501691 | 0.38 |
RARA |
retinoic acid receptor, alpha |
337 |
0.77 |
| chr13_33682757_33682908 | 0.36 |
STARD13-IT1 |
STARD13 intronic transcript 1 (non-protein coding) |
55714 |
0.13 |
| chr2_42795325_42795726 | 0.35 |
MTA3 |
metastasis associated 1 family, member 3 |
132 |
0.97 |
| chr1_156092867_156093791 | 0.35 |
LMNA |
lamin A/C |
2622 |
0.16 |
| chr2_218723094_218723245 | 0.32 |
TNS1 |
tensin 1 |
16286 |
0.25 |
| chr8_49565421_49565933 | 0.31 |
EFCAB1 |
EF-hand calcium binding domain 1 |
76671 |
0.11 |
| chr10_134367045_134367383 | 0.31 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
15571 |
0.21 |
| chr10_72647360_72647920 | 0.31 |
PCBD1 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
901 |
0.61 |
| chr12_2494439_2494710 | 0.30 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
115632 |
0.06 |
| chr7_73868587_73869056 | 0.30 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
382 |
0.88 |
| chr11_65343918_65345038 | 0.30 |
EHBP1L1 |
EH domain binding protein 1-like 1 |
961 |
0.3 |
| chr12_64237243_64237632 | 0.30 |
SRGAP1 |
SLIT-ROBO Rho GTPase activating protein 1 |
636 |
0.66 |
| chr9_130601372_130601801 | 0.28 |
ENSG00000222455 |
. |
1915 |
0.15 |
| chr17_40994290_40994441 | 0.28 |
AOC2 |
amine oxidase, copper containing 2 (retina-specific) |
2252 |
0.13 |
| chr7_128431078_128431544 | 0.28 |
CCDC136 |
coiled-coil domain containing 136 |
153 |
0.93 |
| chrX_39865307_39865929 | 0.27 |
BCOR |
BCL6 corepressor |
56572 |
0.16 |
| chr12_133078846_133079257 | 0.27 |
FBRSL1 |
fibrosin-like 1 |
11894 |
0.2 |
| chr20_2839933_2840363 | 0.27 |
VPS16 |
vacuolar protein sorting 16 homolog (S. cerevisiae) |
2955 |
0.18 |
| chr12_106533043_106533555 | 0.27 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
512 |
0.83 |
| chr1_212405280_212405592 | 0.27 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
53443 |
0.11 |
| chr3_184287134_184287636 | 0.26 |
EPHB3 |
EPH receptor B3 |
7813 |
0.2 |
| chr16_55521708_55521859 | 0.26 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
791 |
0.69 |
| chr10_104436480_104437015 | 0.26 |
TRIM8 |
tripartite motif containing 8 |
32103 |
0.12 |
| chr1_109138261_109138412 | 0.26 |
FAM102B |
family with sequence similarity 102, member B |
35625 |
0.16 |
| chr15_41786624_41787259 | 0.26 |
ITPKA |
inositol-trisphosphate 3-kinase A |
868 |
0.5 |
| chr17_45867102_45867637 | 0.26 |
OSBPL7 |
oxysterol binding protein-like 7 |
31767 |
0.1 |
| chr7_98029982_98030440 | 0.26 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
169 |
0.97 |
| chr8_104152774_104152925 | 0.25 |
BAALC |
brain and acute leukemia, cytoplasmic |
89 |
0.91 |
| chr11_8741654_8741805 | 0.25 |
ST5 |
suppression of tumorigenicity 5 |
1039 |
0.41 |
| chrX_134185283_134186016 | 0.24 |
FAM127B |
family with sequence similarity 127, member B |
556 |
0.74 |
| chr7_1619213_1619508 | 0.24 |
PSMG3 |
proteasome (prosome, macropain) assembly chaperone 3 |
8719 |
0.17 |
| chr8_49289133_49289284 | 0.24 |
ENSG00000252710 |
. |
68618 |
0.14 |
| chr11_3159025_3159707 | 0.23 |
OSBPL5 |
oxysterol binding protein-like 5 |
8807 |
0.17 |
| chr19_55593378_55593915 | 0.23 |
EPS8L1 |
EPS8-like 1 |
1886 |
0.2 |
| chr17_56031434_56032135 | 0.23 |
CUEDC1 |
CUE domain containing 1 |
834 |
0.58 |
| chr9_88899289_88899440 | 0.23 |
ISCA1 |
iron-sulfur cluster assembly 1 |
1688 |
0.36 |
| chr22_50243343_50244155 | 0.23 |
ZBED4 |
zinc finger, BED-type containing 4 |
3741 |
0.22 |
| chr6_56180319_56180615 | 0.23 |
RP3-445N2.1 |
|
16448 |
0.26 |
| chr16_73081442_73081894 | 0.23 |
ZFHX3 |
zinc finger homeobox 3 |
606 |
0.78 |
| chr21_46825889_46826320 | 0.23 |
COL18A1 |
collagen, type XVIII, alpha 1 |
1052 |
0.46 |
| chr16_75284868_75285315 | 0.23 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
246 |
0.89 |
| chr1_38471719_38472149 | 0.23 |
FHL3 |
four and a half LIM domains 3 |
757 |
0.54 |
| chr14_105993130_105993899 | 0.23 |
TMEM121 |
transmembrane protein 121 |
574 |
0.65 |
| chr14_24837858_24838637 | 0.23 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
41 |
0.94 |
| chr1_207980505_207981161 | 0.23 |
ENSG00000203709 |
. |
4965 |
0.27 |
| chr12_117463931_117464197 | 0.22 |
TESC |
tescalcin |
20572 |
0.21 |
| chr6_41513317_41513955 | 0.22 |
RP11-328M4.2 |
|
176 |
0.81 |
| chr16_2518153_2518704 | 0.22 |
RP11-715J22.2 |
|
214 |
0.81 |
| chr8_49341467_49342503 | 0.22 |
ENSG00000252710 |
. |
121395 |
0.06 |
| chr7_5452888_5453131 | 0.22 |
TNRC18 |
trinucleotide repeat containing 18 |
5984 |
0.15 |
| chr17_57432844_57432995 | 0.22 |
YPEL2 |
yippee-like 2 (Drosophila) |
10402 |
0.17 |
| chr2_74777231_74777572 | 0.22 |
LOXL3 |
lysyl oxidase-like 3 |
815 |
0.34 |
| chr14_61747742_61748391 | 0.22 |
TMEM30B |
transmembrane protein 30B |
107 |
0.63 |
| chr16_776196_776865 | 0.22 |
CCDC78 |
coiled-coil domain containing 78 |
57 |
0.81 |
| chr19_1755732_1755984 | 0.22 |
ONECUT3 |
one cut homeobox 3 |
3486 |
0.17 |
| chr3_123509920_123510071 | 0.21 |
MYLK |
myosin light chain kinase |
2693 |
0.3 |
| chr2_242746190_242746341 | 0.21 |
NEU4 |
sialidase 4 |
3655 |
0.12 |
| chr15_41221926_41222903 | 0.21 |
DLL4 |
delta-like 4 (Drosophila) |
823 |
0.5 |
| chr16_28543798_28543949 | 0.21 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
6456 |
0.13 |
| chr16_744329_744818 | 0.21 |
FBXL16 |
F-box and leucine-rich repeat protein 16 |
1452 |
0.14 |
| chr9_131936786_131937499 | 0.21 |
RP11-247A12.8 |
|
944 |
0.4 |
| chr11_1146453_1147024 | 0.21 |
MUC5AC |
mucin 5AC, oligomeric mucus/gel-forming |
4842 |
0.22 |
| chr11_123008436_123008892 | 0.21 |
CTD-2216M2.1 |
|
50846 |
0.11 |
| chr5_157940975_157941126 | 0.20 |
CTD-2363C16.1 |
|
468964 |
0.01 |
| chr1_45308919_45309214 | 0.20 |
PTCH2 |
patched 2 |
331 |
0.79 |
| chr21_46713806_46714461 | 0.20 |
ENSG00000215447 |
. |
6166 |
0.19 |
| chr5_177826208_177826359 | 0.20 |
ENSG00000242341 |
. |
21358 |
0.18 |
| chr11_19791928_19792079 | 0.20 |
NAV2 |
neuron navigator 2 |
7010 |
0.24 |
| chr12_6445245_6445959 | 0.20 |
TNFRSF1A |
tumor necrosis factor receptor superfamily, member 1A |
5412 |
0.13 |
| chr11_125983043_125983210 | 0.20 |
CDON |
cell adhesion associated, oncogene regulated |
49896 |
0.11 |
| chr11_46312622_46312792 | 0.20 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
3970 |
0.21 |
| chr7_75911267_75911842 | 0.20 |
SRRM3 |
serine/arginine repetitive matrix 3 |
378 |
0.84 |
| chr6_21596350_21596989 | 0.20 |
SOX4 |
SRY (sex determining region Y)-box 4 |
2598 |
0.45 |
| chr13_28534615_28535489 | 0.20 |
CDX2 |
caudal type homeobox 2 |
10224 |
0.14 |
| chr19_461630_462007 | 0.20 |
SHC2 |
SHC (Src homology 2 domain containing) transforming protein 2 |
822 |
0.45 |
| chr19_42781870_42782547 | 0.19 |
CIC |
capicua transcriptional repressor |
5964 |
0.09 |
| chr3_123826537_123826688 | 0.19 |
KALRN |
kalirin, RhoGEF kinase |
13054 |
0.2 |
| chr18_34834300_34834543 | 0.19 |
CELF4 |
CUGBP, Elav-like family member 4 |
10296 |
0.2 |
| chr22_24169181_24169332 | 0.19 |
DERL3 |
derlin 3 |
11935 |
0.09 |
| chr16_85784826_85785175 | 0.19 |
C16orf74 |
chromosome 16 open reading frame 74 |
265 |
0.86 |
| chr7_100859763_100860679 | 0.19 |
ZNHIT1 |
zinc finger, HIT-type containing 1 |
728 |
0.34 |
| chr19_44144210_44144820 | 0.19 |
CADM4 |
cell adhesion molecule 4 |
524 |
0.59 |
| chr11_113345070_113345599 | 0.19 |
DRD2 |
dopamine receptor D2 |
508 |
0.8 |
| chr19_13133727_13134698 | 0.19 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
593 |
0.58 |
| chr5_71448848_71449179 | 0.19 |
ENSG00000264099 |
. |
16281 |
0.23 |
| chr11_67141426_67142374 | 0.19 |
CLCF1 |
cardiotrophin-like cytokine factor 1 |
252 |
0.81 |
| chr5_72485513_72485664 | 0.19 |
TMEM174 |
transmembrane protein 174 |
16566 |
0.18 |
| chr22_46525487_46525827 | 0.19 |
ENSG00000207875 |
. |
16091 |
0.1 |
| chr17_40913377_40913932 | 0.19 |
RAMP2 |
receptor (G protein-coupled) activity modifying protein 2 |
378 |
0.71 |
| chr1_156675699_156676565 | 0.19 |
CRABP2 |
cellular retinoic acid binding protein 2 |
524 |
0.6 |
| chr2_12958377_12958528 | 0.19 |
ENSG00000264370 |
. |
80959 |
0.11 |
| chr5_14398322_14398473 | 0.19 |
TRIO |
trio Rho guanine nucleotide exchange factor |
90160 |
0.1 |
| chr2_232419389_232419839 | 0.19 |
NMUR1 |
neuromedin U receptor 1 |
24408 |
0.13 |
| chr17_73992846_73993556 | 0.19 |
CDK3 |
cyclin-dependent kinase 3 |
3786 |
0.12 |
| chr17_38501915_38502668 | 0.18 |
RARA |
retinoic acid receptor, alpha |
798 |
0.45 |
| chr19_19245450_19245992 | 0.18 |
TMEM161A |
transmembrane protein 161A |
2511 |
0.19 |
| chr2_235900449_235900600 | 0.18 |
SH3BP4 |
SH3-domain binding protein 4 |
2960 |
0.42 |
| chr19_2097349_2098074 | 0.18 |
IZUMO4 |
IZUMO family member 4 |
830 |
0.37 |
| chr6_125346782_125347068 | 0.18 |
RNF217 |
ring finger protein 217 |
22433 |
0.24 |
| chr19_13206700_13207253 | 0.18 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
6705 |
0.11 |
| chr5_139049687_139050600 | 0.18 |
CXXC5 |
CXXC finger protein 5 |
4878 |
0.24 |
| chr5_177965097_177965248 | 0.18 |
COL23A1 |
collagen, type XXIII, alpha 1 |
24067 |
0.2 |
| chr17_27459777_27460175 | 0.18 |
MYO18A |
myosin XVIIIA |
7460 |
0.15 |
| chr11_125985239_125985390 | 0.18 |
CDON |
cell adhesion associated, oncogene regulated |
52084 |
0.1 |
| chr19_10928760_10928918 | 0.18 |
ENSG00000207752 |
. |
667 |
0.57 |
| chr3_122803125_122803484 | 0.18 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
17109 |
0.23 |
| chr1_21561803_21561954 | 0.18 |
RP3-329E20.2 |
|
30697 |
0.15 |
| chr22_38850403_38851076 | 0.18 |
KCNJ4 |
potassium inwardly-rectifying channel, subfamily J, member 4 |
466 |
0.74 |
| chr15_67303268_67303419 | 0.18 |
SMAD3 |
SMAD family member 3 |
52758 |
0.16 |
| chr19_1324652_1325370 | 0.18 |
MUM1 |
melanoma associated antigen (mutated) 1 |
29699 |
0.07 |
| chr2_97197247_97197870 | 0.18 |
RP11-363D14.1 |
|
485 |
0.79 |
| chr4_10105956_10106688 | 0.18 |
ENSG00000223086 |
. |
11186 |
0.15 |
| chr17_1534982_1535133 | 0.18 |
SLC43A2 |
solute carrier family 43 (amino acid system L transporter), member 2 |
2927 |
0.14 |
| chr5_179246518_179247261 | 0.18 |
SQSTM1 |
sequestosome 1 |
870 |
0.42 |
| chr5_63932962_63933350 | 0.18 |
FAM159B |
family with sequence similarity 159, member B |
52979 |
0.14 |
| chr19_4115268_4115780 | 0.18 |
MAP2K2 |
mitogen-activated protein kinase kinase 2 |
8590 |
0.13 |
| chr19_54727093_54727479 | 0.18 |
LILRB3 |
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 |
92 |
0.92 |
| chr19_45430034_45430698 | 0.18 |
APOC1P1 |
apolipoprotein C-I pseudogene 1 |
133 |
0.9 |
| chr10_119973133_119973284 | 0.18 |
FAM204A |
family with sequence similarity 204, member A |
128544 |
0.05 |
| chr5_172323723_172324191 | 0.18 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
8263 |
0.19 |
| chr15_90745308_90745813 | 0.18 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
792 |
0.52 |
| chr8_21494888_21495039 | 0.18 |
ENSG00000266630 |
. |
87454 |
0.09 |
| chr21_46347768_46348484 | 0.18 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
507 |
0.62 |
| chr7_15498785_15499062 | 0.18 |
AGMO |
alkylglycerol monooxygenase |
93076 |
0.1 |
| chr1_44883284_44883666 | 0.17 |
RNF220 |
ring finger protein 220 |
5705 |
0.21 |
| chrX_17844639_17844790 | 0.17 |
RAI2 |
retinoic acid induced 2 |
11136 |
0.29 |
| chr6_85845994_85846180 | 0.17 |
NT5E |
5'-nucleotidase, ecto (CD73) |
313722 |
0.01 |
| chr22_50741047_50741266 | 0.17 |
PLXNB2 |
plexin B2 |
1265 |
0.26 |
| chr22_29469276_29469693 | 0.17 |
KREMEN1 |
kringle containing transmembrane protein 1 |
365 |
0.83 |
| chr5_151044309_151044460 | 0.17 |
CTB-113P19.1 |
|
12122 |
0.14 |
| chr19_1361317_1362052 | 0.17 |
MUM1 |
melanoma associated antigen (mutated) 1 |
5321 |
0.1 |
| chr21_37923566_37923717 | 0.17 |
CLDN14 |
claudin 14 |
8743 |
0.23 |
| chr3_141929744_141929895 | 0.17 |
GK5 |
glycerol kinase 5 (putative) |
14609 |
0.22 |
| chr19_54637471_54637622 | 0.17 |
CNOT3 |
CCR4-NOT transcription complex, subunit 3 |
3898 |
0.09 |
| chr12_2560717_2560868 | 0.17 |
CACNA1C-AS3 |
CACNA1C antisense RNA 3 |
155814 |
0.04 |
| chr10_64379208_64379359 | 0.17 |
ZNF365 |
zinc finger protein 365 |
24250 |
0.22 |
| chr13_94725792_94725943 | 0.17 |
ENSG00000212559 |
. |
48015 |
0.19 |
| chr3_72442498_72442652 | 0.17 |
RYBP |
RING1 and YY1 binding protein |
53494 |
0.16 |
| chr17_10128061_10128212 | 0.17 |
GAS7 |
growth arrest-specific 7 |
26268 |
0.18 |
| chr1_54685877_54686028 | 0.17 |
MRPL37 |
mitochondrial ribosomal protein L37 |
10333 |
0.13 |
| chr15_66209053_66209318 | 0.17 |
RAB11A |
RAB11A, member RAS oncogene family |
46834 |
0.13 |
| chr4_3709772_3710024 | 0.17 |
ADRA2C |
adrenoceptor alpha 2C |
58177 |
0.14 |
| chr4_7904697_7904890 | 0.17 |
AFAP1 |
actin filament associated protein 1 |
30753 |
0.17 |
| chr8_118994127_118994278 | 0.17 |
EXT1 |
exostosin glycosyltransferase 1 |
128451 |
0.06 |
| chr2_197189762_197189913 | 0.17 |
ENSG00000264627 |
. |
35704 |
0.16 |
| chr1_211503644_211504001 | 0.17 |
TRAF5 |
TNF receptor-associated factor 5 |
3643 |
0.3 |
| chr19_13213848_13214497 | 0.17 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
197 |
0.89 |
| chr13_114885176_114885502 | 0.17 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
11044 |
0.21 |
| chr5_139104544_139104695 | 0.17 |
ENSG00000200756 |
. |
6944 |
0.22 |
| chr17_40833679_40833973 | 0.16 |
CCR10 |
chemokine (C-C motif) receptor 10 |
35 |
0.91 |
| chr3_170340455_170340606 | 0.16 |
RP11-373E16.3 |
|
33821 |
0.18 |
| chr1_110922648_110923109 | 0.16 |
SLC16A4 |
solute carrier family 16, member 4 |
1073 |
0.36 |
| chr5_72659852_72660019 | 0.16 |
FOXD1 |
forkhead box D1 |
84417 |
0.08 |
| chr19_34311473_34312146 | 0.16 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
13980 |
0.27 |
| chr2_158177617_158177768 | 0.16 |
ERMN |
ermin, ERM-like protein |
4058 |
0.26 |
| chr17_19993225_19993666 | 0.16 |
SPECC1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
3110 |
0.28 |
| chr9_130615331_130616090 | 0.16 |
ENG |
endoglin |
1205 |
0.25 |
| chr9_129961402_129961553 | 0.16 |
RALGPS1 |
Ral GEF with PH domain and SH3 binding motif 1 |
11547 |
0.18 |
| chr5_5350867_5351018 | 0.16 |
KIAA0947 |
KIAA0947 |
71865 |
0.13 |
| chr11_33721547_33721982 | 0.16 |
C11orf91 |
chromosome 11 open reading frame 91 |
179 |
0.94 |
| chr17_76875045_76875748 | 0.16 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
5164 |
0.16 |
| chr11_72470083_72470393 | 0.16 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
116 |
0.94 |
| chr4_10105412_10105563 | 0.16 |
ENSG00000223086 |
. |
12021 |
0.15 |
| chr5_38424510_38424661 | 0.16 |
EGFLAM-AS1 |
EGFLAM antisense RNA 1 |
2893 |
0.23 |
| chr6_24901597_24902091 | 0.16 |
FAM65B |
family with sequence similarity 65, member B |
9351 |
0.22 |
| chr19_16448880_16449487 | 0.16 |
KLF2 |
Kruppel-like factor 2 |
13532 |
0.14 |
| chr11_73039124_73039275 | 0.16 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
16589 |
0.12 |
| chr3_129311849_129312381 | 0.16 |
ENSG00000239437 |
. |
1923 |
0.28 |
| chr1_156096167_156097087 | 0.16 |
LMNA |
lamin A/C |
33 |
0.96 |
| chr17_40492798_40492949 | 0.16 |
STAT5A |
signal transducer and activator of transcription 5A |
34685 |
0.09 |
| chr7_492646_493023 | 0.16 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
65311 |
0.11 |
| chr16_4378324_4378965 | 0.16 |
GLIS2 |
GLIS family zinc finger 2 |
3581 |
0.16 |
| chr17_66348034_66348185 | 0.16 |
ARSG |
arylsulfatase G |
60450 |
0.09 |
| chr13_24205823_24205974 | 0.16 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
52379 |
0.16 |
| chr20_50028653_50029381 | 0.16 |
ENSG00000263645 |
. |
35159 |
0.18 |
| chr17_38128356_38128623 | 0.16 |
GSDMA |
gasdermin A |
6713 |
0.12 |
| chr5_31854978_31855350 | 0.16 |
ENSG00000251887 |
. |
22618 |
0.13 |
| chr1_22745301_22745452 | 0.16 |
ZBTB40 |
zinc finger and BTB domain containing 40 |
32968 |
0.19 |
| chr4_1237782_1238175 | 0.16 |
CTBP1 |
C-terminal binding protein 1 |
2267 |
0.22 |
| chr8_93031342_93031493 | 0.16 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
14 |
0.99 |
| chr12_12016346_12017014 | 0.16 |
ETV6 |
ets variant 6 |
22191 |
0.26 |
| chr2_47201847_47202176 | 0.16 |
RP11-15I20.1 |
|
17984 |
0.15 |
| chr15_74906801_74907591 | 0.16 |
CLK3 |
CDC-like kinase 3 |
26 |
0.93 |
| chr3_87003421_87003572 | 0.16 |
VGLL3 |
vestigial like 3 (Drosophila) |
36356 |
0.24 |
| chr12_63161016_63161167 | 0.16 |
ENSG00000200296 |
. |
83590 |
0.09 |
| chr8_144922026_144922763 | 0.16 |
NRBP2 |
nuclear receptor binding protein 2 |
408 |
0.7 |
| chr5_175299080_175299718 | 0.16 |
CPLX2 |
complexin 2 |
414 |
0.87 |
| chr10_3493784_3493935 | 0.16 |
PITRM1 |
pitrilysin metallopeptidase 1 |
278856 |
0.01 |
| chr3_45127227_45127378 | 0.16 |
ENSG00000252410 |
. |
51222 |
0.11 |
| chr17_36862672_36863533 | 0.15 |
CTB-58E17.3 |
|
588 |
0.44 |
| chr18_32656273_32656556 | 0.15 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
34800 |
0.22 |
| chr11_43701160_43701492 | 0.15 |
HSD17B12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
933 |
0.68 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.1 | 0.4 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.1 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0051892 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.0 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0022009 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0044320 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.0 | GO:2000300 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.5 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.0 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.0 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) negative regulation of cation transmembrane transport(GO:1904063) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.0 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0071675 | positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:2000649 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.0 | GO:0051138 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.0 | GO:0006168 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.0 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.0 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.0 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0030146 | obsolete diuresis(GO:0030146) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.0 | GO:1903670 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.0 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.0 | GO:2000258 | negative regulation of humoral immune response(GO:0002921) negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.0 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.0 | GO:0043205 | fibril(GO:0043205) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.0 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.0 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.0 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |