Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
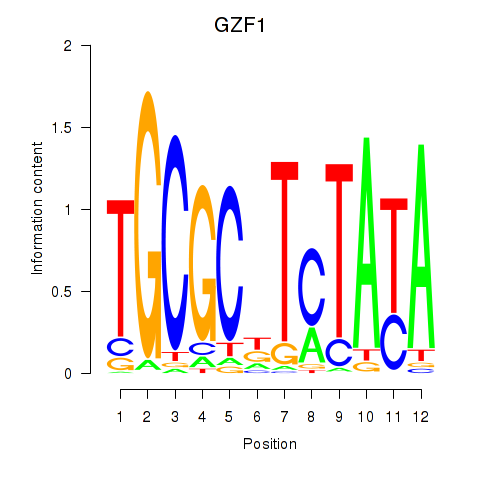
Results for GZF1
Z-value: 0.99
Transcription factors associated with GZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GZF1
|
ENSG00000125812.11 | GDNF inducible zinc finger protein 1 |
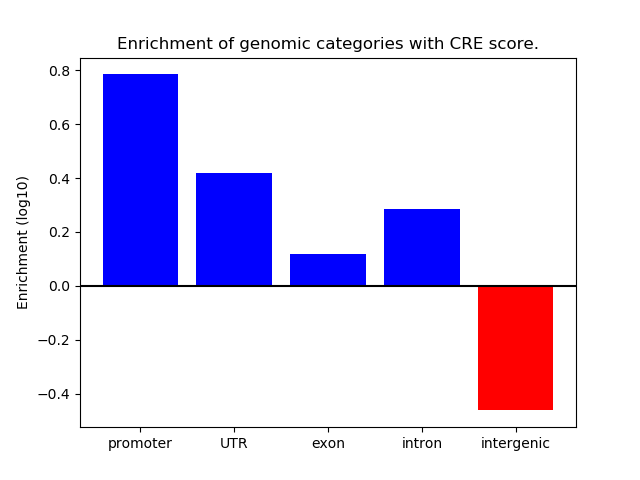
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr20_23346727_23346946 | GZF1 | 1815 | 0.212392 | 0.85 | 4.0e-03 | Click! |
| chr20_23346451_23346602 | GZF1 | 1505 | 0.253265 | 0.75 | 2.0e-02 | Click! |
| chr20_23348740_23348891 | GZF1 | 3794 | 0.130752 | -0.63 | 6.7e-02 | Click! |
| chr20_23346043_23346194 | GZF1 | 1097 | 0.347683 | 0.60 | 9.0e-02 | Click! |
| chr20_23346955_23347106 | GZF1 | 2009 | 0.194471 | 0.54 | 1.3e-01 | Click! |
Activity of the GZF1 motif across conditions
Conditions sorted by the z-value of the GZF1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
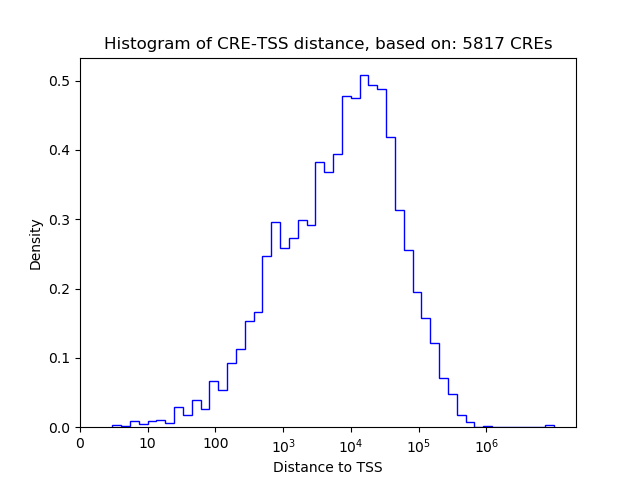
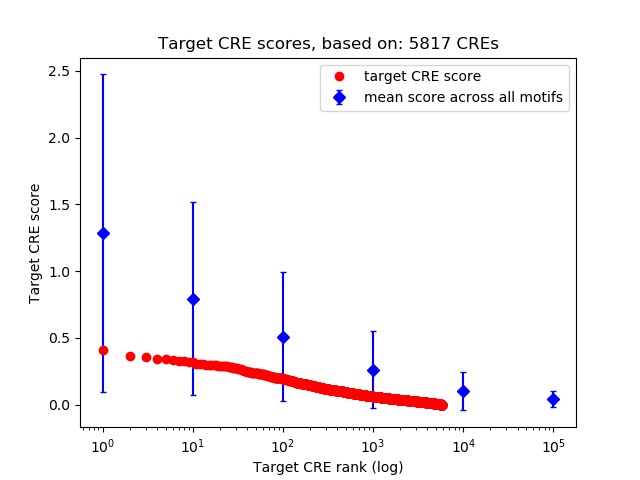
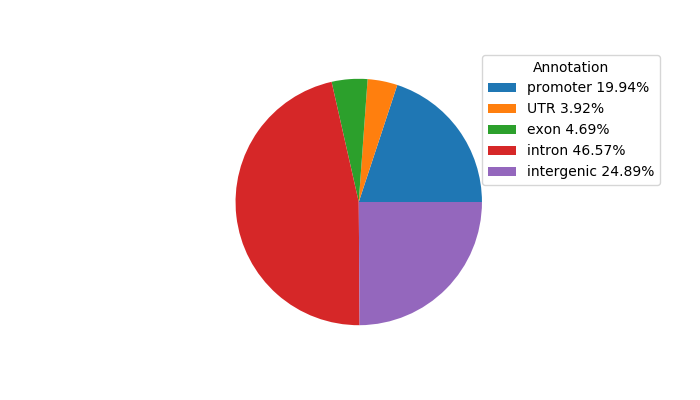
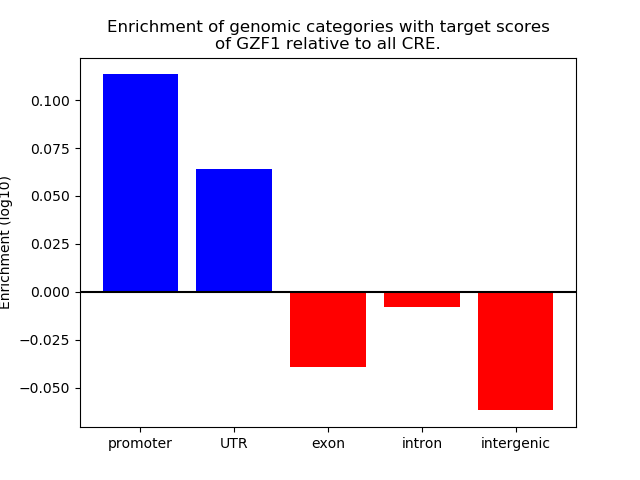
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_140058363_140058567 | 0.41 |
SLC37A3 |
solute carrier family 37, member 3 |
6565 |
0.15 |
| chr11_67049148_67049384 | 0.37 |
ANKRD13D |
ankyrin repeat domain 13 family, member D |
6752 |
0.12 |
| chr14_59950418_59950569 | 0.35 |
L3HYPDH |
L-3-hydroxyproline dehydratase (trans-) |
90 |
0.91 |
| chr12_129278613_129278971 | 0.35 |
SLC15A4 |
solute carrier family 15 (oligopeptide transporter), member 4 |
7454 |
0.27 |
| chr5_157286748_157286966 | 0.35 |
CLINT1 |
clathrin interactor 1 |
674 |
0.75 |
| chrX_153534844_153535009 | 0.34 |
TKTL1 |
transketolase-like 1 |
1517 |
0.24 |
| chr21_15855177_15855351 | 0.33 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
63398 |
0.12 |
| chr14_36789095_36789307 | 0.32 |
MBIP |
MAP3K12 binding inhibitory protein 1 |
125 |
0.97 |
| chr6_35838535_35838686 | 0.32 |
SRPK1 |
SRSF protein kinase 1 |
50214 |
0.11 |
| chr2_198119149_198119485 | 0.32 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
47926 |
0.12 |
| chr1_168504102_168504301 | 0.30 |
XCL2 |
chemokine (C motif) ligand 2 |
9034 |
0.25 |
| chrX_65081944_65082241 | 0.30 |
RP6-159A1.3 |
|
137501 |
0.05 |
| chr19_54928713_54928864 | 0.30 |
TTYH1 |
tweety family member 1 |
2088 |
0.17 |
| chr20_34077500_34077678 | 0.30 |
RP3-477O4.14 |
|
1154 |
0.34 |
| chr20_48417718_48417888 | 0.30 |
ENSG00000252540 |
. |
6455 |
0.17 |
| chr7_38256301_38256485 | 0.29 |
STARD3NL |
STARD3 N-terminal like |
38396 |
0.22 |
| chr2_187351499_187351878 | 0.29 |
ZC3H15 |
zinc finger CCCH-type containing 15 |
708 |
0.69 |
| chr8_126718847_126719004 | 0.29 |
ENSG00000206695 |
. |
194270 |
0.03 |
| chr2_62427282_62427433 | 0.29 |
B3GNT2 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
4109 |
0.2 |
| chr10_73499941_73500092 | 0.29 |
C10orf105 |
chromosome 10 open reading frame 105 |
2435 |
0.3 |
| chr16_50309553_50309704 | 0.29 |
ADCY7 |
adenylate cyclase 7 |
1564 |
0.41 |
| chr17_68285276_68285427 | 0.29 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
119675 |
0.06 |
| chr9_132651876_132652490 | 0.29 |
FNBP1 |
formin binding protein 1 |
29406 |
0.13 |
| chr2_113940950_113941232 | 0.29 |
AC016683.5 |
|
8154 |
0.14 |
| chr10_30863847_30863998 | 0.28 |
ENSG00000239744 |
. |
19089 |
0.22 |
| chr16_22218333_22218484 | 0.28 |
EEF2K |
eukaryotic elongation factor-2 kinase |
805 |
0.63 |
| chr11_111957057_111957216 | 0.28 |
SDHD |
succinate dehydrogenase complex, subunit D, integral membrane protein |
361 |
0.46 |
| chr10_118652800_118653001 | 0.28 |
ENO4 |
enolase family member 4 |
34355 |
0.17 |
| chr11_118055454_118055605 | 0.28 |
SCN2B |
sodium channel, voltage-gated, type II, beta subunit |
8141 |
0.15 |
| chr22_40322306_40322457 | 0.27 |
GRAP2 |
GRB2-related adaptor protein 2 |
214 |
0.93 |
| chr4_109103898_109104112 | 0.27 |
ENSG00000232021 |
. |
6561 |
0.25 |
| chr10_104005871_104006203 | 0.27 |
GBF1 |
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
748 |
0.56 |
| chr2_2000070_2000423 | 0.27 |
MYT1L |
myelin transcription factor 1-like |
153588 |
0.04 |
| chr1_77748105_77748256 | 0.26 |
AK5 |
adenylate kinase 5 |
107 |
0.98 |
| chr14_107130545_107130696 | 0.26 |
IGHV3-66 |
immunoglobulin heavy variable 3-66 |
940 |
0.18 |
| chr20_419998_420268 | 0.26 |
ENSG00000206797 |
. |
11174 |
0.15 |
| chr14_50557550_50557701 | 0.25 |
C14orf183 |
chromosome 14 open reading frame 183 |
1736 |
0.29 |
| chr1_239550914_239551159 | 0.25 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
241337 |
0.02 |
| chr6_15315532_15315775 | 0.25 |
ENSG00000201367 |
. |
502 |
0.81 |
| chr1_230296504_230296655 | 0.24 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
93561 |
0.08 |
| chr5_130405277_130405629 | 0.24 |
HINT1 |
histidine triad nucleotide binding protein 1 |
92867 |
0.09 |
| chr2_46665870_46666021 | 0.24 |
ENSG00000241791 |
. |
9705 |
0.2 |
| chr3_87040067_87040263 | 0.24 |
VGLL3 |
vestigial like 3 (Drosophila) |
92 |
0.99 |
| chr5_118668072_118668246 | 0.24 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
711 |
0.71 |
| chr7_102938360_102938511 | 0.24 |
PMPCB |
peptidase (mitochondrial processing) beta |
532 |
0.76 |
| chr1_38315162_38315404 | 0.24 |
MTF1 |
metal-regulatory transcription factor 1 |
10009 |
0.1 |
| chr11_749652_749928 | 0.24 |
TALDO1 |
transaldolase 1 |
2362 |
0.12 |
| chr6_11374826_11375046 | 0.24 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
7596 |
0.29 |
| chr1_145928317_145928496 | 0.24 |
PDZK1P1 |
PDZ domain containing 1 pseudogene 1 |
11707 |
0.16 |
| chr1_160577265_160577416 | 0.24 |
CD84 |
CD84 molecule |
28034 |
0.12 |
| chrX_142720411_142720668 | 0.24 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
1780 |
0.48 |
| chr20_25370250_25370403 | 0.24 |
ABHD12 |
abhydrolase domain containing 12 |
444 |
0.85 |
| chr17_3816161_3816362 | 0.23 |
P2RX1 |
purinergic receptor P2X, ligand-gated ion channel, 1 |
3533 |
0.2 |
| chr22_32560080_32560278 | 0.23 |
C22orf42 |
chromosome 22 open reading frame 42 |
4904 |
0.17 |
| chr17_2696074_2696428 | 0.23 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
3481 |
0.22 |
| chr1_110047130_110047281 | 0.23 |
AMIGO1 |
adhesion molecule with Ig-like domain 1 |
5099 |
0.11 |
| chr11_95094119_95094342 | 0.23 |
ENSG00000201204 |
. |
112308 |
0.06 |
| chr7_6422686_6422957 | 0.23 |
RAC1 |
ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
8651 |
0.18 |
| chr15_64720549_64720703 | 0.23 |
TRIP4 |
thyroid hormone receptor interactor 4 |
4312 |
0.13 |
| chrX_75392533_75392684 | 0.23 |
PBDC1 |
polysaccharide biosynthesis domain containing 1 |
163 |
0.97 |
| chr14_101292334_101292485 | 0.23 |
AL117190.2 |
|
3128 |
0.07 |
| chr10_14615009_14615187 | 0.23 |
FAM107B |
family with sequence similarity 107, member B |
760 |
0.73 |
| chr1_54843922_54844304 | 0.22 |
SSBP3 |
single stranded DNA binding protein 3 |
27064 |
0.19 |
| chr8_99956309_99956579 | 0.22 |
OSR2 |
odd-skipped related transciption factor 2 |
226 |
0.9 |
| chrX_19868073_19868317 | 0.22 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
37382 |
0.2 |
| chr9_134879882_134880105 | 0.22 |
MED27 |
mediator complex subunit 27 |
75260 |
0.11 |
| chr3_112059886_112060037 | 0.22 |
CD200 |
CD200 molecule |
7936 |
0.25 |
| chr6_1850485_1850731 | 0.22 |
FOXC1 |
forkhead box C1 |
239927 |
0.02 |
| chr9_7977771_7977988 | 0.22 |
TMEM261 |
transmembrane protein 261 |
177812 |
0.04 |
| chr12_32115294_32115445 | 0.21 |
KIAA1551 |
KIAA1551 |
54 |
0.98 |
| chr11_2812322_2812544 | 0.21 |
KCNQ1-AS1 |
KCNQ1 antisense RNA 1 |
70365 |
0.08 |
| chr9_131133838_131134081 | 0.21 |
URM1 |
ubiquitin related modifier 1 |
309 |
0.81 |
| chr1_205583719_205584005 | 0.21 |
ELK4 |
ELK4, ETS-domain protein (SRF accessory protein 1) |
17228 |
0.14 |
| chr17_56407049_56407274 | 0.21 |
BZRAP1-AS1 |
BZRAP1 antisense RNA 1 |
195 |
0.84 |
| chr12_116986256_116986469 | 0.21 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
10824 |
0.28 |
| chr21_15916640_15916923 | 0.21 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
1881 |
0.42 |
| chr3_32344273_32344503 | 0.21 |
ENSG00000207857 |
. |
43168 |
0.14 |
| chr3_179675139_179675303 | 0.21 |
PEX5L |
peroxisomal biogenesis factor 5-like |
15819 |
0.24 |
| chr9_74390054_74390240 | 0.21 |
TMEM2 |
transmembrane protein 2 |
6347 |
0.31 |
| chr20_24271413_24271883 | 0.20 |
ENSG00000200231 |
. |
53757 |
0.17 |
| chr1_110611840_110612052 | 0.20 |
ALX3 |
ALX homeobox 3 |
1376 |
0.31 |
| chr16_17478039_17478236 | 0.20 |
XYLT1 |
xylosyltransferase I |
86601 |
0.11 |
| chr17_78247850_78248001 | 0.20 |
RNF213 |
ring finger protein 213 |
13256 |
0.15 |
| chr20_48439855_48440008 | 0.20 |
ENSG00000252123 |
. |
4598 |
0.18 |
| chr1_147470017_147470168 | 0.20 |
ENSG00000206791 |
. |
16723 |
0.19 |
| chr6_106545393_106545544 | 0.20 |
PRDM1 |
PR domain containing 1, with ZNF domain |
1013 |
0.56 |
| chr14_50319739_50319890 | 0.20 |
NEMF |
nuclear export mediator factor |
107 |
0.89 |
| chr9_97090440_97090723 | 0.20 |
NUTM2F |
NUT family member 2F |
345 |
0.89 |
| chr11_110708426_110708577 | 0.20 |
ARHGAP20 |
Rho GTPase activating protein 20 |
124589 |
0.06 |
| chr8_6566732_6567036 | 0.20 |
AGPAT5 |
1-acylglycerol-3-phosphate O-acyltransferase 5 |
670 |
0.58 |
| chr9_138881295_138881528 | 0.20 |
UBAC1 |
UBA domain containing 1 |
28185 |
0.17 |
| chr11_36423240_36423525 | 0.20 |
PRR5L |
proline rich 5 like |
767 |
0.66 |
| chr9_95877175_95877370 | 0.20 |
C9orf89 |
chromosome 9 open reading frame 89 |
18772 |
0.14 |
| chr11_72322937_72323189 | 0.20 |
ENSG00000272036 |
. |
3111 |
0.17 |
| chr22_46684505_46684828 | 0.20 |
GTSE1 |
G-2 and S-phase expressed 1 |
7972 |
0.15 |
| chr2_8517146_8517394 | 0.20 |
AC011747.7 |
|
298626 |
0.01 |
| chr2_2000524_2000698 | 0.20 |
MYT1L |
myelin transcription factor 1-like |
153953 |
0.04 |
| chr2_134913656_134913807 | 0.20 |
ENSG00000263813 |
. |
29035 |
0.2 |
| chr9_99691569_99691774 | 0.20 |
NUTM2G |
NUT family member 2G |
385 |
0.89 |
| chr8_21916870_21917081 | 0.20 |
DMTN |
dematin actin binding protein |
241 |
0.88 |
| chr3_45018010_45018161 | 0.20 |
EXOSC7 |
exosome component 7 |
363 |
0.51 |
| chr2_203776193_203776517 | 0.19 |
CARF |
calcium responsive transcription factor |
582 |
0.45 |
| chr7_129714164_129714315 | 0.19 |
KLHDC10 |
kelch domain containing 10 |
3627 |
0.19 |
| chr3_5233671_5233989 | 0.19 |
EDEM1 |
ER degradation enhancer, mannosidase alpha-like 1 |
3796 |
0.18 |
| chrX_19758311_19758804 | 0.19 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
398 |
0.92 |
| chr3_171197799_171197950 | 0.19 |
TNIK |
TRAF2 and NCK interacting kinase |
19677 |
0.19 |
| chr20_1473982_1474133 | 0.19 |
SIRPB2 |
signal-regulatory protein beta 2 |
1930 |
0.25 |
| chr2_232165655_232165809 | 0.19 |
ARMC9 |
armadillo repeat containing 9 |
30487 |
0.16 |
| chr2_143908158_143909031 | 0.19 |
RP11-190J23.1 |
|
21147 |
0.22 |
| chr22_37319301_37319452 | 0.19 |
CSF2RB |
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
423 |
0.77 |
| chr2_86080450_86080721 | 0.19 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
14192 |
0.17 |
| chr6_35558805_35559048 | 0.19 |
ENSG00000212579 |
. |
60669 |
0.09 |
| chr2_143883334_143883719 | 0.19 |
ARHGAP15 |
Rho GTPase activating protein 15 |
3357 |
0.33 |
| chr7_99965278_99965466 | 0.18 |
PILRA |
paired immunoglobin-like type 2 receptor alpha |
219 |
0.85 |
| chr21_36411020_36411357 | 0.18 |
RUNX1 |
runt-related transcription factor 1 |
10274 |
0.32 |
| chr4_129835794_129835945 | 0.18 |
JADE1 |
jade family PHD finger 1 |
83281 |
0.11 |
| chr9_77728037_77728188 | 0.18 |
OSTF1 |
osteoclast stimulating factor 1 |
24653 |
0.16 |
| chr8_11558200_11558351 | 0.18 |
GATA4 |
GATA binding protein 4 |
3385 |
0.25 |
| chr15_41688710_41688871 | 0.18 |
NDUFAF1 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
5850 |
0.16 |
| chr11_2502582_2502749 | 0.18 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
19548 |
0.16 |
| chr6_159074020_159074298 | 0.18 |
SYTL3 |
synaptotagmin-like 3 |
3113 |
0.24 |
| chr1_212463226_212463384 | 0.18 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
4426 |
0.19 |
| chr17_25808384_25808701 | 0.18 |
KSR1 |
kinase suppressor of ras 1 |
9506 |
0.2 |
| chr2_40712943_40713094 | 0.18 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
26483 |
0.27 |
| chr17_4391049_4391200 | 0.18 |
RP13-580F15.2 |
|
1476 |
0.3 |
| chr2_201827346_201827560 | 0.18 |
ORC2 |
origin recognition complex, subunit 2 |
528 |
0.42 |
| chr15_29247145_29247302 | 0.18 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
6284 |
0.25 |
| chr6_112311435_112311966 | 0.18 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
63575 |
0.11 |
| chr6_13434535_13434686 | 0.18 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
26241 |
0.16 |
| chr14_65165014_65165274 | 0.18 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
5676 |
0.24 |
| chr19_10449056_10449486 | 0.18 |
ICAM3 |
intercellular adhesion molecule 3 |
1024 |
0.31 |
| chr2_2000849_2001005 | 0.17 |
MYT1L |
myelin transcription factor 1-like |
154269 |
0.04 |
| chr11_121366062_121366254 | 0.17 |
RP11-730K11.1 |
|
42436 |
0.18 |
| chr11_121205690_121205841 | 0.17 |
SC5D |
sterol-C5-desaturase |
42204 |
0.19 |
| chr14_25457947_25458098 | 0.17 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
21824 |
0.27 |
| chr18_61642862_61643013 | 0.17 |
SERPINB8 |
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
4105 |
0.22 |
| chrX_48796553_48796937 | 0.17 |
OTUD5 |
OTU domain containing 5 |
17706 |
0.08 |
| chr9_87905756_87905907 | 0.17 |
AGTPBP1 |
ATP/GTP binding protein 1 |
390370 |
0.01 |
| chr2_43397843_43397994 | 0.17 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
55830 |
0.13 |
| chr5_39204147_39204382 | 0.17 |
FYB |
FYN binding protein |
1135 |
0.63 |
| chr21_34672110_34672341 | 0.17 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
24509 |
0.13 |
| chr2_103059024_103059175 | 0.16 |
AC007278.3 |
|
3926 |
0.17 |
| chr1_36835365_36835563 | 0.16 |
STK40 |
serine/threonine kinase 40 |
8523 |
0.13 |
| chr1_27951892_27952043 | 0.16 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
679 |
0.61 |
| chr14_92413353_92413602 | 0.16 |
FBLN5 |
fibulin 5 |
59 |
0.98 |
| chr7_45019149_45019431 | 0.16 |
MYO1G |
myosin IG |
593 |
0.65 |
| chr12_125473857_125474087 | 0.16 |
DHX37 |
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
305 |
0.9 |
| chr15_64665673_64665824 | 0.16 |
CTD-2116N17.1 |
Uncharacterized protein |
163 |
0.92 |
| chr20_44833333_44833580 | 0.16 |
CDH22 |
cadherin 22, type 2 |
46878 |
0.14 |
| chr20_10485213_10485441 | 0.16 |
SLX4IP |
SLX4 interacting protein |
69376 |
0.11 |
| chrX_135826724_135826994 | 0.16 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
22643 |
0.16 |
| chr5_77224775_77224926 | 0.16 |
TBCA |
tubulin folding cofactor A |
60246 |
0.14 |
| chr3_182990531_182990717 | 0.16 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
7492 |
0.21 |
| chr7_148406333_148406484 | 0.16 |
CUL1 |
cullin 1 |
10404 |
0.18 |
| chr10_26729391_26729542 | 0.16 |
APBB1IP |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
2112 |
0.43 |
| chr6_166582383_166582539 | 0.16 |
T |
T, brachyury homolog (mouse) |
273 |
0.93 |
| chr16_88751434_88751599 | 0.16 |
SNAI3-AS1 |
SNAI3 antisense RNA 1 |
25 |
0.93 |
| chr10_101288694_101289161 | 0.16 |
RP11-129J12.2 |
|
1727 |
0.34 |
| chr6_147091501_147091674 | 0.16 |
ADGB |
androglobin |
6 |
0.99 |
| chr6_128842055_128842211 | 0.16 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
263 |
0.91 |
| chr6_36818115_36818266 | 0.16 |
CPNE5 |
copine V |
10412 |
0.17 |
| chr1_27683765_27684109 | 0.16 |
MAP3K6 |
mitogen-activated protein kinase kinase kinase 6 |
975 |
0.41 |
| chr15_99332143_99332351 | 0.16 |
ENSG00000264480 |
. |
4592 |
0.3 |
| chr6_74230738_74230889 | 0.16 |
EEF1A1 |
eukaryotic translation elongation factor 1 alpha 1 |
72 |
0.95 |
| chr18_60988556_60988796 | 0.16 |
BCL2 |
B-cell CLL/lymphoma 2 |
1315 |
0.41 |
| chr6_53205632_53205819 | 0.16 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
7862 |
0.22 |
| chr6_10656054_10656229 | 0.16 |
ENSG00000201048 |
. |
18860 |
0.13 |
| chr17_43488233_43488428 | 0.16 |
ARHGAP27 |
Rho GTPase activating protein 27 |
538 |
0.66 |
| chr6_142987864_142988015 | 0.16 |
RP1-67K17.3 |
|
81641 |
0.11 |
| chr1_231893166_231893324 | 0.16 |
ENSG00000222986 |
. |
86864 |
0.09 |
| chr11_48055026_48055177 | 0.16 |
AC103828.1 |
|
17694 |
0.2 |
| chr7_8218098_8218600 | 0.15 |
ENSG00000265212 |
. |
23697 |
0.19 |
| chr6_108109017_108109337 | 0.15 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
15722 |
0.26 |
| chr21_43612105_43612375 | 0.15 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
7559 |
0.2 |
| chr3_187675103_187675412 | 0.15 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
195815 |
0.03 |
| chr20_5093353_5093504 | 0.15 |
TMEM230 |
transmembrane protein 230 |
29 |
0.95 |
| chr10_50380716_50380985 | 0.15 |
C10orf128 |
chromosome 10 open reading frame 128 |
15507 |
0.17 |
| chr5_67581048_67581199 | 0.15 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
3073 |
0.38 |
| chr15_41245906_41246096 | 0.15 |
CHAC1 |
ChaC, cation transport regulator homolog 1 (E. coli) |
403 |
0.79 |
| chr16_57703759_57703957 | 0.15 |
GPR97 |
G protein-coupled receptor 97 |
1638 |
0.28 |
| chr16_30461637_30461788 | 0.15 |
ENSG00000202476 |
. |
3706 |
0.08 |
| chr20_31139955_31140106 | 0.15 |
RP11-410N8.3 |
|
8865 |
0.16 |
| chr22_31619679_31619930 | 0.15 |
ENSG00000199695 |
. |
965 |
0.4 |
| chr17_62067658_62067809 | 0.15 |
C17orf72 |
chromosome 17 open reading frame 72 |
7978 |
0.14 |
| chr1_167509501_167509771 | 0.15 |
CREG1 |
cellular repressor of E1A-stimulated genes 1 |
13368 |
0.19 |
| chr19_6668717_6669176 | 0.15 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
1182 |
0.4 |
| chr20_24026643_24026819 | 0.15 |
GGTLC1 |
gamma-glutamyltransferase light chain 1 |
57315 |
0.15 |
| chr7_92219738_92219889 | 0.15 |
FAM133B |
family with sequence similarity 133, member B |
105 |
0.97 |
| chr2_151344116_151344336 | 0.15 |
RND3 |
Rho family GTPase 3 |
17 |
0.99 |
| chr1_77513564_77513779 | 0.15 |
RP4-564M11.2 |
|
19934 |
0.23 |
| chr8_22929951_22930102 | 0.15 |
RP11-875O11.3 |
|
1975 |
0.21 |
| chr21_43319494_43319645 | 0.15 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
2192 |
0.23 |
| chr18_60202275_60202426 | 0.15 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
4397 |
0.31 |
| chr2_135008118_135008541 | 0.15 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
3501 |
0.31 |
| chr10_71108085_71108264 | 0.15 |
ENSG00000221774 |
. |
15223 |
0.17 |
| chr7_38262255_38262771 | 0.15 |
STARD3NL |
STARD3 N-terminal like |
44516 |
0.2 |
| chr15_65567487_65567698 | 0.15 |
ENSG00000206903 |
. |
10337 |
0.12 |
| chr12_9140443_9140771 | 0.15 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
1530 |
0.35 |
| chr13_73614878_73615077 | 0.15 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
14137 |
0.24 |
| chr12_9144662_9144957 | 0.15 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
143 |
0.95 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.1 | 0.3 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.1 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.2 | GO:0034227 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.2 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.2 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:1904376 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.3 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.0 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0061054 | dermatome development(GO:0061054) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.0 | 0.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:0071333 | cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:0097300 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.0 | GO:0071637 | heterotypic cell-cell adhesion(GO:0034113) regulation of heterotypic cell-cell adhesion(GO:0034114) monocyte chemotactic protein-1 production(GO:0071605) regulation of monocyte chemotactic protein-1 production(GO:0071637) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0002674 | negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0031420 | alkali metal ion binding(GO:0031420) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | Genes involved in Metabolism of lipids and lipoproteins |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.0 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.0 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |