Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
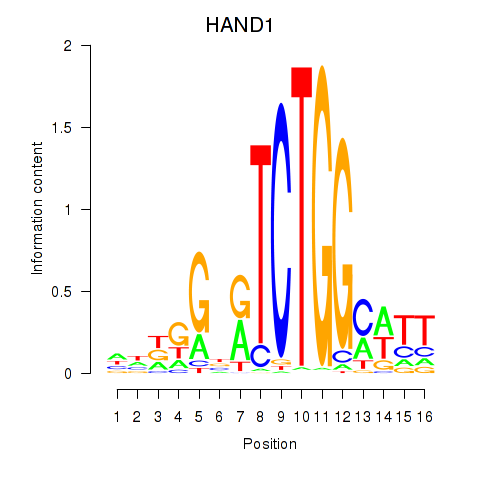
Results for HAND1
Z-value: 0.99
Transcription factors associated with HAND1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HAND1
|
ENSG00000113196.2 | heart and neural crest derivatives expressed 1 |
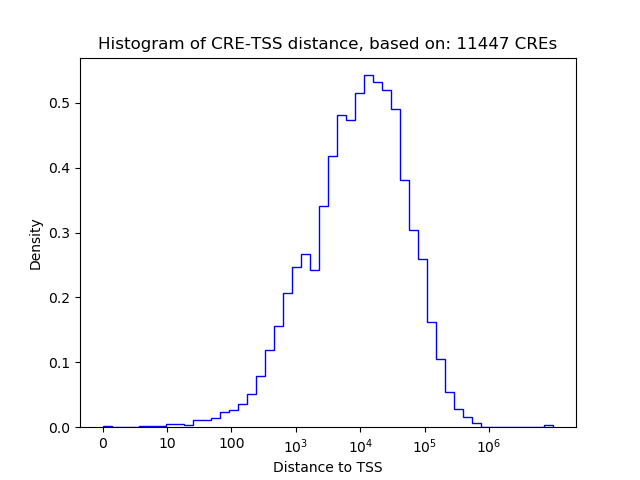
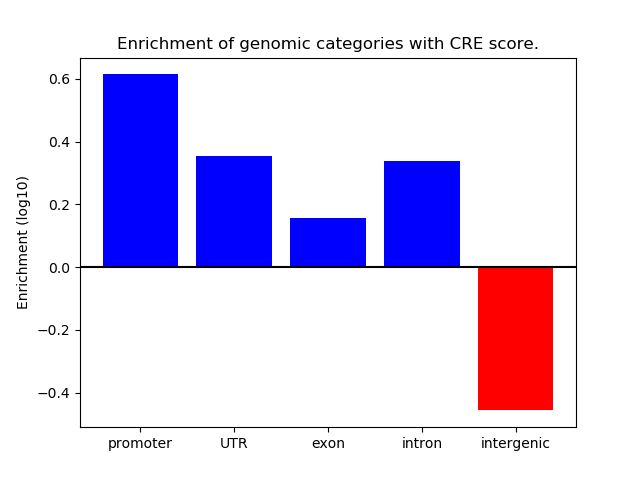
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_153900655_153900951 | HAND1 | 42979 | 0.133708 | -0.73 | 2.6e-02 | Click! |
| chr5_153856378_153856529 | HAND1 | 1371 | 0.419329 | 0.72 | 2.8e-02 | Click! |
| chr5_153853814_153853984 | HAND1 | 3925 | 0.213488 | 0.55 | 1.2e-01 | Click! |
| chr5_153852765_153852916 | HAND1 | 4984 | 0.197080 | 0.53 | 1.5e-01 | Click! |
| chr5_153845026_153845177 | HAND1 | 12723 | 0.160774 | -0.49 | 1.8e-01 | Click! |
Activity of the HAND1 motif across conditions
Conditions sorted by the z-value of the HAND1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
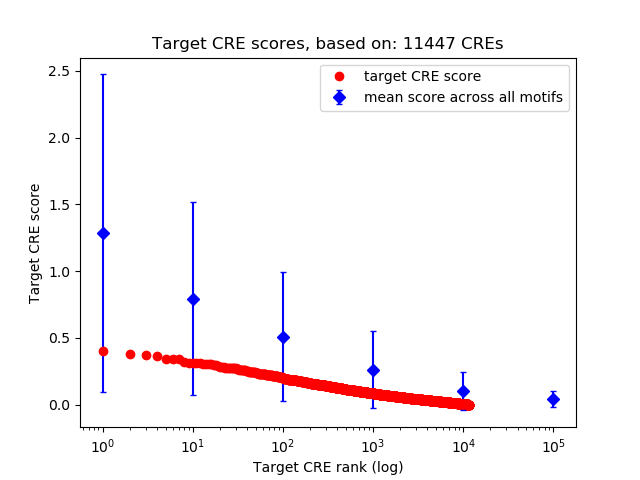
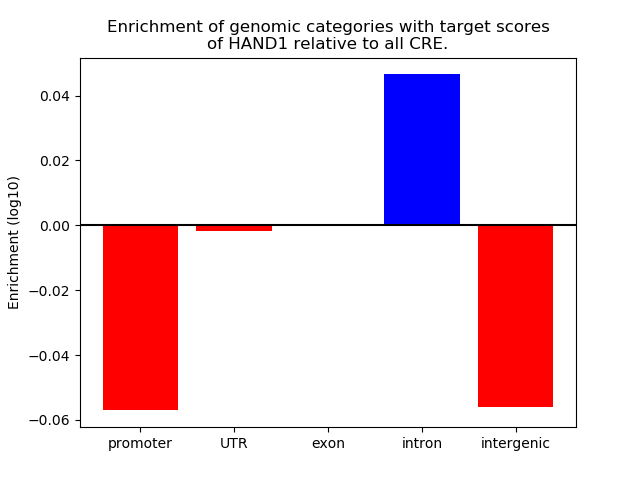
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_97009046_97009425 | 0.40 |
NCAPH |
non-SMC condensin I complex, subunit H |
7679 |
0.15 |
| chr17_37252182_37252943 | 0.38 |
PLXDC1 |
plexin domain containing 1 |
11178 |
0.15 |
| chr13_114892713_114893126 | 0.37 |
RASA3 |
RAS p21 protein activator 3 |
5108 |
0.24 |
| chr11_4111133_4111394 | 0.36 |
RRM1 |
ribonucleotide reductase M1 |
4776 |
0.22 |
| chr14_67948872_67949023 | 0.34 |
TMEM229B |
transmembrane protein 229B |
6502 |
0.18 |
| chrX_47482564_47483099 | 0.34 |
SYN1 |
synapsin I |
3579 |
0.15 |
| chr4_15769645_15769984 | 0.34 |
CD38 |
CD38 molecule |
10087 |
0.23 |
| chr1_65342372_65342702 | 0.32 |
JAK1 |
Janus kinase 1 |
89650 |
0.08 |
| chr1_28296104_28296455 | 0.31 |
RP11-460I13.2 |
|
9575 |
0.13 |
| chr3_33472190_33472341 | 0.31 |
UBP1 |
upstream binding protein 1 (LBP-1a) |
9237 |
0.21 |
| chr8_93029450_93029601 | 0.31 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
66 |
0.98 |
| chr12_104973749_104974059 | 0.31 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
8722 |
0.24 |
| chr2_127418703_127419020 | 0.31 |
GYPC |
glycophorin C (Gerbich blood group) |
5101 |
0.3 |
| chr5_14439836_14439987 | 0.31 |
TRIO |
trio Rho guanine nucleotide exchange factor |
48646 |
0.18 |
| chr3_11677644_11677795 | 0.31 |
VGLL4 |
vestigial like 4 (Drosophila) |
7679 |
0.22 |
| chr6_126142988_126143327 | 0.31 |
NCOA7-AS1 |
NCOA7 antisense RNA 1 |
3153 |
0.24 |
| chr1_198503645_198504115 | 0.30 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
5924 |
0.32 |
| chr3_113949300_113949543 | 0.29 |
ZNF80 |
zinc finger protein 80 |
7004 |
0.18 |
| chr13_40762290_40762441 | 0.29 |
ENSG00000207458 |
. |
38599 |
0.22 |
| chr1_27076332_27076532 | 0.28 |
ENSG00000266138 |
. |
13683 |
0.13 |
| chr12_68176752_68176973 | 0.28 |
RP11-335O4.3 |
|
112163 |
0.07 |
| chr16_67836589_67836775 | 0.28 |
RANBP10 |
RAN binding protein 10 |
3778 |
0.12 |
| chr19_2775327_2775584 | 0.28 |
SGTA |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
4676 |
0.12 |
| chr1_19768716_19768889 | 0.28 |
ENSG00000240490 |
. |
17924 |
0.15 |
| chr2_136578827_136578978 | 0.28 |
AC011893.3 |
|
1141 |
0.45 |
| chr1_90064313_90064509 | 0.28 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
30611 |
0.14 |
| chr1_40137074_40137225 | 0.28 |
NT5C1A |
5'-nucleotidase, cytosolic IA |
561 |
0.66 |
| chr1_221957617_221957791 | 0.27 |
DUSP10 |
dual specificity phosphatase 10 |
42186 |
0.19 |
| chr1_221508301_221508621 | 0.27 |
DUSP10 |
dual specificity phosphatase 10 |
402341 |
0.01 |
| chr6_112044961_112045268 | 0.27 |
FYN |
FYN oncogene related to SRC, FGR, YES |
3849 |
0.3 |
| chr10_74923079_74923230 | 0.27 |
ECD |
ecdysoneless homolog (Drosophila) |
4663 |
0.12 |
| chr6_109036108_109036376 | 0.27 |
FOXO3 |
forkhead box O3 |
58693 |
0.15 |
| chr11_128396911_128397217 | 0.26 |
RP11-1007G5.2 |
|
1027 |
0.59 |
| chr14_98675980_98676131 | 0.26 |
ENSG00000222066 |
. |
122032 |
0.06 |
| chr10_121044351_121044502 | 0.26 |
RP11-79M19.2 |
|
51268 |
0.11 |
| chr17_34040072_34040441 | 0.26 |
RASL10B |
RAS-like, family 10, member B |
18412 |
0.11 |
| chr7_8167446_8167940 | 0.26 |
AC006042.6 |
|
14038 |
0.21 |
| chr3_16944411_16944593 | 0.26 |
PLCL2 |
phospholipase C-like 2 |
18050 |
0.23 |
| chr2_47170482_47170698 | 0.25 |
MCFD2 |
multiple coagulation factor deficiency 2 |
1596 |
0.28 |
| chr11_104670330_104670481 | 0.25 |
CASP12 |
caspase 12 (gene/pseudogene) |
98736 |
0.08 |
| chr13_110383641_110384047 | 0.25 |
LINC00676 |
long intergenic non-protein coding RNA 676 |
3215 |
0.35 |
| chr2_98582630_98582781 | 0.25 |
TMEM131 |
transmembrane protein 131 |
29649 |
0.2 |
| chr10_63840836_63841344 | 0.25 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
32120 |
0.19 |
| chr7_132116532_132116686 | 0.25 |
AC011625.1 |
|
79516 |
0.11 |
| chr17_17084049_17084292 | 0.25 |
MPRIP |
myosin phosphatase Rho interacting protein |
1298 |
0.29 |
| chr19_654179_654330 | 0.25 |
RNF126 |
ring finger protein 126 |
7148 |
0.09 |
| chr1_229889719_229889932 | 0.24 |
URB2 |
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
103231 |
0.07 |
| chr2_38925814_38925965 | 0.24 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
17452 |
0.16 |
| chr9_123679203_123679400 | 0.24 |
TRAF1 |
TNF receptor-associated factor 1 |
2451 |
0.31 |
| chr11_60755747_60755977 | 0.24 |
ENSG00000207153 |
. |
3223 |
0.16 |
| chr1_54744952_54745103 | 0.24 |
RP5-997D24.3 |
|
6051 |
0.18 |
| chr3_187897306_187897937 | 0.23 |
AC022498.1 |
Uncharacterized protein |
1290 |
0.5 |
| chrX_44125263_44125482 | 0.23 |
EFHC2 |
EF-hand domain (C-terminal) containing 2 |
77546 |
0.11 |
| chr14_61984119_61984270 | 0.23 |
RP11-47I22.4 |
|
11652 |
0.2 |
| chr7_2178117_2178407 | 0.23 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
29026 |
0.21 |
| chr3_134385868_134386019 | 0.23 |
KY |
kyphoscoliosis peptidase |
16079 |
0.23 |
| chr14_92341016_92341177 | 0.23 |
FBLN5 |
fibulin 5 |
3290 |
0.26 |
| chr7_640540_641424 | 0.23 |
PRKAR1B |
protein kinase, cAMP-dependent, regulatory, type I, beta |
1338 |
0.36 |
| chrX_19778652_19778939 | 0.23 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
12974 |
0.29 |
| chr4_73932257_73932427 | 0.23 |
COX18 |
COX18 cytochrome C oxidase assembly factor |
3067 |
0.32 |
| chr2_30488933_30489313 | 0.23 |
LBH |
limb bud and heart development |
34077 |
0.19 |
| chr9_131832251_131832515 | 0.23 |
DOLPP1 |
dolichyldiphosphatase 1 |
10996 |
0.11 |
| chr8_125649609_125649792 | 0.23 |
RP11-532M24.1 |
|
57828 |
0.12 |
| chr2_99323973_99324148 | 0.23 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
19099 |
0.21 |
| chr2_175644427_175644584 | 0.23 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
15305 |
0.23 |
| chr1_207098582_207098736 | 0.23 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
2067 |
0.25 |
| chr3_10284470_10284734 | 0.22 |
TATDN2 |
TatD DNase domain containing 2 |
5105 |
0.14 |
| chr5_110590081_110590232 | 0.22 |
AC010468.2 |
|
15631 |
0.2 |
| chr6_12163632_12164063 | 0.22 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
37868 |
0.22 |
| chr2_45029072_45029470 | 0.22 |
ENSG00000252896 |
. |
20002 |
0.27 |
| chr3_15359672_15359823 | 0.22 |
ENSG00000238891 |
. |
1620 |
0.31 |
| chr2_109790847_109791252 | 0.22 |
ENSG00000264934 |
. |
33005 |
0.2 |
| chr16_16135222_16135434 | 0.22 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
31615 |
0.19 |
| chr1_206752443_206752919 | 0.22 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
22188 |
0.14 |
| chr2_12644304_12644548 | 0.22 |
ENSG00000207183 |
. |
92799 |
0.09 |
| chr8_12860265_12860557 | 0.22 |
ENSG00000206996 |
. |
41448 |
0.16 |
| chr17_26134829_26135084 | 0.22 |
NOS2 |
nitric oxide synthase 2, inducible |
7431 |
0.21 |
| chr9_132033309_132033483 | 0.22 |
ENSG00000220992 |
. |
40400 |
0.13 |
| chr17_75452925_75453139 | 0.22 |
SEPT9 |
septin 9 |
584 |
0.7 |
| chr11_128290296_128290447 | 0.21 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
84918 |
0.1 |
| chr14_104383228_104383519 | 0.21 |
C14orf2 |
chromosome 14 open reading frame 2 |
4458 |
0.17 |
| chr3_15578936_15579113 | 0.21 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
15766 |
0.14 |
| chr4_90600499_90600650 | 0.21 |
RP11-115D19.1 |
|
2089 |
0.47 |
| chr17_14106066_14106394 | 0.21 |
AC005224.2 |
|
7575 |
0.21 |
| chr11_68272006_68272464 | 0.21 |
ENSG00000222339 |
. |
884 |
0.48 |
| chr2_144302669_144302857 | 0.21 |
RP11-570L15.2 |
|
26911 |
0.19 |
| chr10_15212970_15213280 | 0.21 |
NMT2 |
N-myristoyltransferase 2 |
2433 |
0.32 |
| chr3_38539403_38539554 | 0.21 |
EXOG |
endo/exonuclease (5'-3'), endonuclease G-like |
1506 |
0.38 |
| chrX_78420820_78421003 | 0.21 |
GPR174 |
G protein-coupled receptor 174 |
5558 |
0.35 |
| chr18_2702393_2702544 | 0.21 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
46582 |
0.11 |
| chrX_19766528_19766693 | 0.21 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
789 |
0.77 |
| chr3_177661794_177662251 | 0.21 |
ENSG00000199858 |
. |
63545 |
0.15 |
| chr2_190149930_190150081 | 0.21 |
ENSG00000266817 |
. |
26482 |
0.23 |
| chr4_48344741_48344971 | 0.21 |
SLAIN2 |
SLAIN motif family, member 2 |
1517 |
0.49 |
| chr11_65870833_65870984 | 0.20 |
PACS1 |
phosphofurin acidic cluster sorting protein 1 |
2674 |
0.17 |
| chr3_119732053_119732204 | 0.20 |
GSK3B |
glycogen synthase kinase 3 beta |
80385 |
0.09 |
| chr17_75960444_75960595 | 0.20 |
TNRC6C |
trinucleotide repeat containing 6C |
39730 |
0.14 |
| chr12_52654824_52655351 | 0.20 |
KRT121P |
keratin 121 pseudogene |
2436 |
0.15 |
| chr1_32667214_32667874 | 0.20 |
CCDC28B |
coiled-coil domain containing 28B |
1310 |
0.22 |
| chr11_69519758_69519936 | 0.20 |
FGF19 |
fibroblast growth factor 19 |
437 |
0.84 |
| chr8_71580114_71580327 | 0.20 |
LACTB2 |
lactamase, beta 2 |
1172 |
0.34 |
| chr3_16865405_16865955 | 0.20 |
PLCL2 |
phospholipase C-like 2 |
60772 |
0.14 |
| chr22_37226429_37226580 | 0.20 |
PVALB |
parvalbumin |
10981 |
0.13 |
| chr2_144301655_144301806 | 0.20 |
RP11-570L15.2 |
|
27944 |
0.19 |
| chr15_65574741_65574985 | 0.20 |
ENSG00000206903 |
. |
3066 |
0.17 |
| chr1_167604982_167605729 | 0.20 |
RP3-455J7.4 |
|
5444 |
0.21 |
| chr9_139298829_139298980 | 0.20 |
SDCCAG3 |
serologically defined colon cancer antigen 3 |
2733 |
0.14 |
| chr5_34570275_34570426 | 0.19 |
RAI14 |
retinoic acid induced 14 |
85992 |
0.09 |
| chr10_35808142_35808293 | 0.19 |
GJD4 |
gap junction protein, delta 4, 40.1kDa |
86121 |
0.08 |
| chr10_127824965_127825297 | 0.19 |
ENSG00000222740 |
. |
9020 |
0.29 |
| chr14_98677293_98677464 | 0.19 |
ENSG00000222066 |
. |
120709 |
0.06 |
| chr7_47791048_47791199 | 0.19 |
LINC00525 |
long intergenic non-protein coding RNA 525 |
9951 |
0.23 |
| chr10_111776497_111776648 | 0.19 |
ADD3 |
adducin 3 (gamma) |
8850 |
0.21 |
| chr10_11358280_11358431 | 0.19 |
CELF2-AS1 |
CELF2 antisense RNA 1 |
3492 |
0.36 |
| chr4_6919752_6920098 | 0.19 |
TBC1D14 |
TBC1 domain family, member 14 |
7950 |
0.19 |
| chr19_7581365_7581909 | 0.19 |
ZNF358 |
zinc finger protein 358 |
633 |
0.51 |
| chr20_39775264_39775450 | 0.19 |
RP1-1J6.2 |
|
8714 |
0.2 |
| chr12_104977859_104978010 | 0.19 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
4692 |
0.26 |
| chr9_18385263_18385414 | 0.19 |
ADAMTSL1 |
ADAMTS-like 1 |
88554 |
0.1 |
| chr12_111893200_111893523 | 0.19 |
ATXN2 |
ataxin 2 |
555 |
0.76 |
| chr14_62543804_62543955 | 0.19 |
RP11-355I22.7 |
|
33714 |
0.23 |
| chr15_101303814_101303965 | 0.19 |
ALDH1A3 |
aldehyde dehydrogenase 1 family, member A3 |
114030 |
0.05 |
| chr16_29988360_29988511 | 0.19 |
TAOK2 |
TAO kinase 2 |
2833 |
0.11 |
| chr6_37471487_37471725 | 0.19 |
CCDC167 |
coiled-coil domain containing 167 |
3908 |
0.25 |
| chr2_42332803_42333174 | 0.18 |
PKDCC |
protein kinase domain containing, cytoplasmic |
57828 |
0.11 |
| chr10_105208792_105209256 | 0.18 |
RP11-225H22.7 |
|
929 |
0.37 |
| chr8_8704312_8704638 | 0.18 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
46680 |
0.14 |
| chr14_98909850_98910089 | 0.18 |
ENSG00000241757 |
. |
91968 |
0.09 |
| chr12_12016346_12017014 | 0.18 |
ETV6 |
ets variant 6 |
22191 |
0.26 |
| chr7_134512686_134512837 | 0.18 |
CALD1 |
caldesmon 1 |
15874 |
0.27 |
| chr3_43359417_43359828 | 0.18 |
ENSG00000241939 |
. |
3179 |
0.23 |
| chr11_118567796_118568096 | 0.18 |
ENSG00000207462 |
. |
5105 |
0.14 |
| chr1_32730988_32731377 | 0.18 |
LCK |
lymphocyte-specific protein tyrosine kinase |
8532 |
0.09 |
| chr9_137285314_137285572 | 0.18 |
RXRA |
retinoid X receptor, alpha |
12985 |
0.22 |
| chr7_105537147_105537368 | 0.18 |
ATXN7L1 |
ataxin 7-like 1 |
20207 |
0.25 |
| chr15_48931540_48931691 | 0.18 |
FBN1 |
fibrillin 1 |
6303 |
0.29 |
| chr14_72908650_72908927 | 0.18 |
RGS6 |
regulator of G-protein signaling 6 |
12440 |
0.31 |
| chr15_70196429_70196590 | 0.18 |
ENSG00000215958 |
. |
118666 |
0.06 |
| chr8_117744312_117744463 | 0.18 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
23636 |
0.2 |
| chr19_44436594_44436745 | 0.18 |
ZNF45 |
zinc finger protein 45 |
2742 |
0.17 |
| chr10_112431690_112431897 | 0.18 |
RBM20 |
RNA binding motif protein 20 |
27638 |
0.14 |
| chr20_55036334_55036485 | 0.18 |
ENSG00000212084 |
. |
996 |
0.44 |
| chr7_24600789_24600940 | 0.18 |
MPP6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
12023 |
0.24 |
| chr15_42068094_42068245 | 0.18 |
MAPKBP1 |
mitogen-activated protein kinase binding protein 1 |
1269 |
0.33 |
| chr15_100782496_100782647 | 0.18 |
ADAMTS17 |
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
98770 |
0.07 |
| chr2_143898363_143898928 | 0.18 |
ARHGAP15 |
Rho GTPase activating protein 15 |
11762 |
0.25 |
| chr11_587585_588264 | 0.18 |
PHRF1 |
PHD and ring finger domains 1 |
11403 |
0.07 |
| chr1_25800396_25800562 | 0.18 |
TMEM57 |
transmembrane protein 57 |
43068 |
0.11 |
| chr2_225810689_225810885 | 0.18 |
DOCK10 |
dedicator of cytokinesis 10 |
995 |
0.69 |
| chr12_76945362_76945546 | 0.18 |
OSBPL8 |
oxysterol binding protein-like 8 |
7830 |
0.32 |
| chr6_45705684_45705835 | 0.18 |
ENSG00000252738 |
. |
91918 |
0.09 |
| chrY_22739941_22740092 | 0.18 |
EIF1AY |
eukaryotic translation initiation factor 1A, Y-linked |
2336 |
0.46 |
| chrX_64901581_64901971 | 0.18 |
MSN |
moesin |
14239 |
0.29 |
| chr1_90126896_90127047 | 0.18 |
LRRC8C |
leucine rich repeat containing 8 family, member C |
28340 |
0.15 |
| chr5_171579294_171579519 | 0.18 |
STK10 |
serine/threonine kinase 10 |
35984 |
0.15 |
| chr8_126946234_126946440 | 0.18 |
ENSG00000206695 |
. |
33142 |
0.25 |
| chr2_28624680_28624911 | 0.18 |
RP11-373D23.2 |
|
5113 |
0.18 |
| chr14_22972883_22973034 | 0.18 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
16787 |
0.09 |
| chr17_64371368_64371623 | 0.18 |
ENSG00000244044 |
. |
11348 |
0.22 |
| chr11_75040450_75040782 | 0.17 |
ENSG00000199090 |
. |
5614 |
0.16 |
| chr2_235388644_235388795 | 0.17 |
ARL4C |
ADP-ribosylation factor-like 4C |
16525 |
0.3 |
| chr1_161167348_161167499 | 0.17 |
ADAMTS4 |
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
1420 |
0.16 |
| chr1_198713615_198713766 | 0.17 |
RP11-553K8.5 |
|
77500 |
0.1 |
| chr4_122087583_122087734 | 0.17 |
TNIP3 |
TNFAIP3 interacting protein 3 |
2163 |
0.36 |
| chr22_24868016_24868188 | 0.17 |
UPB1 |
ureidopropionase, beta |
4896 |
0.18 |
| chr14_103249481_103249632 | 0.17 |
TRAF3 |
TNF receptor-associated factor 3 |
5601 |
0.2 |
| chrX_100877410_100877579 | 0.17 |
ARMCX3 |
armadillo repeat containing, X-linked 3 |
293 |
0.8 |
| chr14_99647460_99647611 | 0.17 |
AL162151.4 |
|
22782 |
0.23 |
| chr18_9090620_9090967 | 0.17 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
11835 |
0.18 |
| chr12_15791050_15791201 | 0.17 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
24083 |
0.2 |
| chr10_104022395_104022570 | 0.17 |
ENSG00000251989 |
. |
1495 |
0.33 |
| chr20_44509042_44509193 | 0.17 |
ZSWIM1 |
zinc finger, SWIM-type containing 1 |
749 |
0.41 |
| chr20_2722949_2723333 | 0.17 |
EBF4 |
early B-cell factor 4 |
36254 |
0.09 |
| chr20_18523568_18523719 | 0.17 |
SEC23B |
Sec23 homolog B (S. cerevisiae) |
631 |
0.61 |
| chr5_133478997_133479191 | 0.17 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
611 |
0.75 |
| chr4_10621970_10622121 | 0.17 |
CLNK |
cytokine-dependent hematopoietic cell linker |
36140 |
0.23 |
| chr1_233416939_233417108 | 0.17 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
14436 |
0.26 |
| chrX_68051807_68052101 | 0.17 |
EFNB1 |
ephrin-B1 |
3114 |
0.39 |
| chr4_108978798_108979031 | 0.17 |
HADH |
hydroxyacyl-CoA dehydrogenase |
53123 |
0.13 |
| chr6_35278431_35279308 | 0.17 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
1354 |
0.42 |
| chr9_92043695_92043846 | 0.17 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
7608 |
0.26 |
| chr15_33321925_33322076 | 0.17 |
FMN1 |
formin 1 |
38085 |
0.18 |
| chr6_24893842_24894069 | 0.17 |
FAM65B |
family with sequence similarity 65, member B |
16372 |
0.2 |
| chr1_167472608_167472887 | 0.17 |
CD247 |
CD247 molecule |
15028 |
0.19 |
| chr2_113938283_113938445 | 0.17 |
AC016683.5 |
|
5427 |
0.15 |
| chr5_130613594_130613745 | 0.17 |
CDC42SE2 |
CDC42 small effector 2 |
13876 |
0.28 |
| chr17_42085806_42085957 | 0.17 |
NAGS |
N-acetylglutamate synthase |
3756 |
0.12 |
| chr1_224570147_224570348 | 0.16 |
ENSG00000266618 |
. |
15766 |
0.14 |
| chr1_109048430_109048671 | 0.16 |
FAM102B |
family with sequence similarity 102, member B |
54161 |
0.13 |
| chr19_8676942_8677296 | 0.16 |
ADAMTS10 |
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
1534 |
0.33 |
| chr6_139467698_139467849 | 0.16 |
HECA |
headcase homolog (Drosophila) |
11524 |
0.25 |
| chr18_36157629_36157780 | 0.16 |
ENSG00000222704 |
. |
438604 |
0.01 |
| chr14_69950322_69950473 | 0.16 |
PLEKHD1 |
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
1074 |
0.6 |
| chr15_66152500_66152651 | 0.16 |
RAB11A |
RAB11A, member RAS oncogene family |
9219 |
0.19 |
| chr1_52258333_52258484 | 0.16 |
RP4-657D16.3 |
|
1229 |
0.38 |
| chr9_123665604_123665920 | 0.16 |
TRAF1 |
TNF receptor-associated factor 1 |
11088 |
0.19 |
| chr6_156342570_156342721 | 0.16 |
ENSG00000221456 |
. |
74714 |
0.13 |
| chr14_71276715_71276967 | 0.16 |
MAP3K9 |
mitogen-activated protein kinase kinase kinase 9 |
590 |
0.84 |
| chr1_157083117_157083268 | 0.16 |
ETV3L |
ets variant 3-like |
13592 |
0.19 |
| chr20_25022537_25022783 | 0.16 |
ACSS1 |
acyl-CoA synthetase short-chain family member 1 |
9321 |
0.2 |
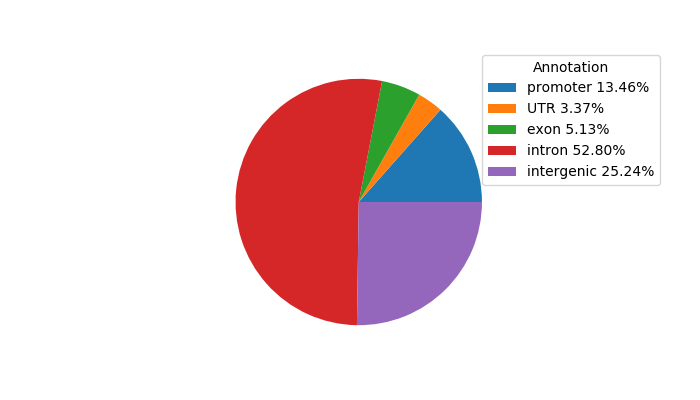
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.1 | 0.2 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.3 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0051892 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.1 | GO:2000351 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.0 | GO:0002251 | organ or tissue specific immune response(GO:0002251) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.3 | GO:0051806 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.5 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.0 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.0 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.0 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.0 | GO:0002604 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.0 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.1 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0051194 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of nucleoside metabolic process(GO:0045979) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) positive regulation of ATP metabolic process(GO:1903580) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0033084 | regulation of immature T cell proliferation in thymus(GO:0033084) |
| 0.0 | 0.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.0 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.3 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.0 | GO:1902339 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.0 | GO:0032691 | negative regulation of interleukin-1 beta production(GO:0032691) negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.0 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.0 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.0 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway(GO:0033145) |
| 0.0 | 0.1 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 0.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0002577 | regulation of antigen processing and presentation(GO:0002577) positive regulation of antigen processing and presentation(GO:0002579) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0042533 | tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.0 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.0 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.0 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |